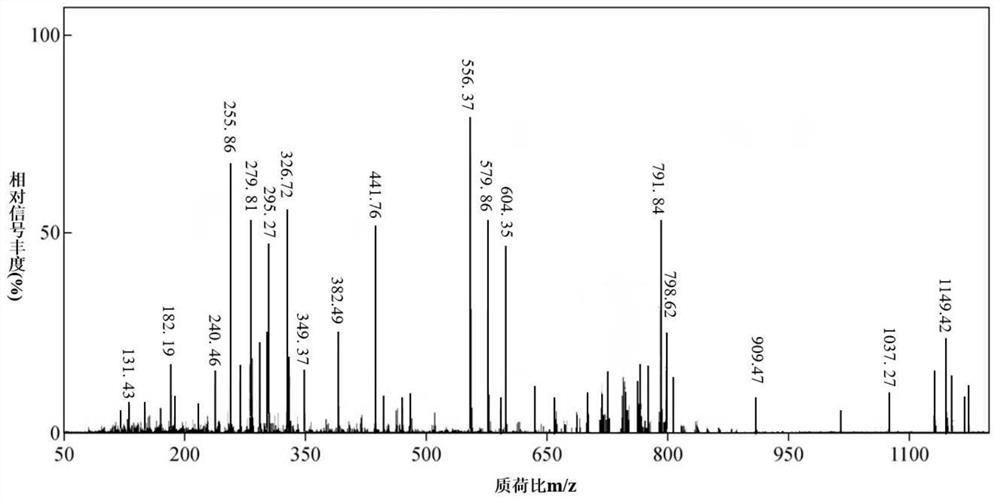
Detection method of lipid content in biological liver tissue
A technology of liver tissue and detection method, which is applied to measurement devices, instruments, scientific instruments, etc., can solve the problems of poor repeatability, low sensitivity, interference in the detection process of lipid content, etc., and achieves improvement of lipid extraction rate and accuracy. , Taking into account the effect of lipid extraction rate and impurity removal rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0041] In step S3, the preparation method of the filler includes the following steps:
[0042] S31, adding silica gel particles into the hydrochloric acid solution, fully mixing at 90-100° C. for 14-16 hours, filtering, washing and drying to obtain activated silica gel;
[0043] S32. Add methylimidazole and 3-chloropropyltriethoxysilane into the acetonitrile solution, react at 100-110°C for 16-20 hours, then add potassium hexafluorophosphate, continue the reaction for 24-28 hours, filter and Collect the filtrate;
[0044] S33. After mixing the filtrate obtained in step S32 with toluene, add the activated silica gel obtained in step S31, react at 100-110°C for 22-26 hours, filter, wash and dry the product to obtain functionalization silica gel;
[0045] S34. Add the functionalized silica gel obtained in step S33 into the acetone solution of silver tetrafluoroborate, mix thoroughly for 8-10 hours under the condition of avoiding light, filter, wash and dry to obtain silica gel ...
Embodiment 1
[0051] This embodiment provides a method for detecting lipid content in biological liver tissue, comprising the following steps:
[0052] S1. Homogenization and primary extraction
[0053] Add 100mg of mouse liver tissue into the homogenization tube containing ceramic microbeads, then add 5mL of the first mixed solvent made of isopropanol and acetonitrile at a volume ratio of 1:2.5, and homogenize at a speed of 3000r / min for 15s , and the homogenate was obtained after repeating the homogenization twice.
[0054] After continuing to centrifuge the homogenate at a speed of 3000 r / min for 10 min, take the supernatant as the first extract.
[0055] S2, secondary extraction
[0056] Take 500 μL of the first extract obtained in step S1, add 750 μL of the second mixed solvent formed by mixing n-hexane and ethyl acetate at a volume ratio of 3.5:1, and then add 750 μL of 1.5% aqueous acetic acid solution, After vortexing for 20 s, centrifuge at 3000 r / min for 10 min, and take the su...
Embodiment 2~9 and comparative example 1~2
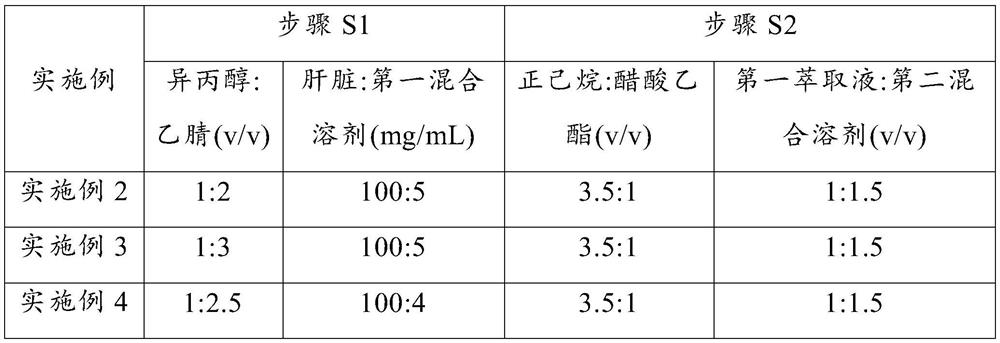
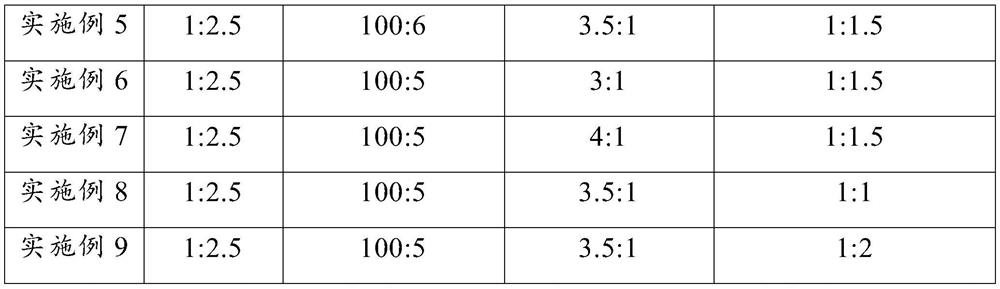
[0074] Embodiments 2 to 9 respectively provide a detection method for the lipid content of biological liver tissue. Compared with embodiment 1, the difference is that some parameters in steps S1 to S2 are changed. The detection parameters corresponding to each embodiment are as follows: As shown in Table 2, the rest of the steps are consistent with those in Example 1, and will not be repeated here.
[0075] The corresponding parameter of each step in the embodiment 2~9 of table 2
[0076]
[0077]
[0078] Comparative Example 1 and Comparative Example 2 respectively provide a detection method for the lipid content of biological liver tissue. Compared with Example 1, the difference is that in step S1 of Comparative Example 1, deionized water is used instead of the first mixed solvent to carry out Detection; step S2 was not performed in comparative example 2, and the first extract obtained in step S1 was directly subjected to solid-phase extraction; the rest of the steps i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com