African swine fever virus P22 protein nanoparticle and preparation method and application thereof
An African swine fever virus and nanoparticle technology, applied in the field of African swine fever virus P22 protein nanoparticles and their preparation, can solve the problems of inability to resist virulent attack, low protective effect, and no protective effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1 plasmid construction
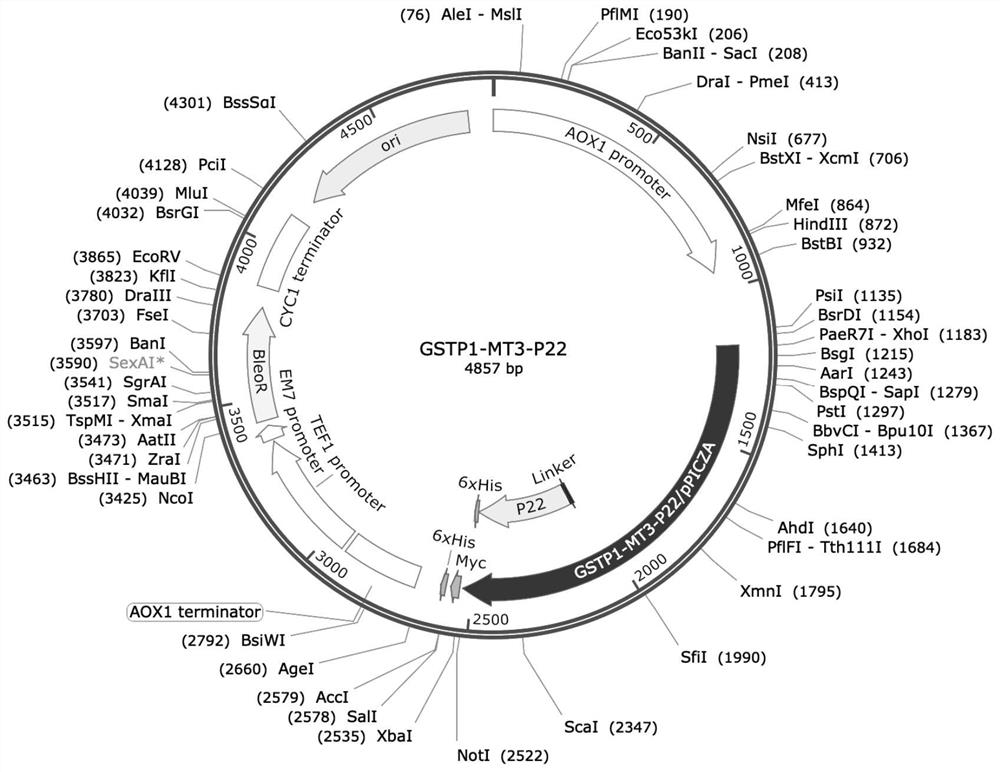
[0043] This embodiment provides the amino acid sequence of African swine fever virus P22 protein nanoparticle (GSTP1-MT3-P22), as shown in SEQ ID NO:4. Entrust Qingke Biological Company to synthesize the DNA fragment of GSTP1-MT3-P22 with His tag fused at the C-terminus, its nucleic acid sequence is shown in SEQ ID NO: 5 (among them, the 13th-1317th bits encode the DNA fragment shown in SEQ ID NO: 4 sequence, the 1318-1335th encoding His tag), and constructed into the plasmid vector pPICZaA, the map of the constructed plasmid is shown in figure 1 As indicated, the plasmid was named pPICZaA-GSTP1-MT3-P22. The plasmid vector pPICZaA contains the α-factor secretion signal peptide sequence before inserting the fragment. The sequenced correct monoclonal strain was cultivated, and the plasmid pPICZaA-GSTP1-MT3-P22 was extracted for use.
[0044] Amino acid sequence of GSTP1-MT3-P22 (SEQ ID NO: 4):
[0045] DPETCPCPSGGSCTCADSCKCEGCKCTSCK...
Embodiment 2
[0046] Example 2 Induced expression of protein nanoparticles
[0047] After the plasmid prepared in Example 1 was linearized, it was electroporated into X-33 yeast competent cells for inducible expression. The specific steps are as follows:
[0048] (1) Plasmid linearization
[0049] The plasmid pPICZaA-GSTP1-MT3-P22 prepared in Example 1 was taken and digested with linearizing enzyme SacI, and the restriction system was as follows:
[0050]
[0051] After mixing, centrifuge at 12000 rpm at room temperature for 1 min to shake the liquid to the bottom of the tube, and bathe in water at 37 °C for 4 h.
[0052] After digestion, take 2 μL, and conduct 1% agarose gel electrophoresis with the original plasmid to identify the linearization result. After identification, the plasmid has been digested completely. Use the plasmid kit to recover the digested plasmid and use it in the subsequent test steps. .
[0053] (2) Electroporation (aseptic operation)
[0054] Add 10μL of the ...
Embodiment 3
[0058] Example 3 SDS-PAGE identification
[0059] Take 1 mL of the culture product prepared in Example 2, centrifuge in a centrifuge at the maximum speed for 2 min at room temperature, take the supernatant, mix the supernatant with Loading buffer to make a loading sample, carry out SDS-PAGE identification, and analyze the expression level. SDS-PAGE identification results see figure 2 As shown in (left), it was identified that the target protein was successfully expressed.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com