Pathogen molecule detection method based on nanopore sequencing
A nanopore sequencing and molecular detection technology, applied in the biological field, can solve the problems of low sensitivity, long detection time, time-consuming PCR, etc., and achieve the effects of high accuracy/consistency, fast sequencing speed, and low detection limit.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0049] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0050] Aiming at the problems existing in the prior art, the present invention provides a method for detecting pathogenic molecules based on nanopore sequencing. The present invention will be described in detail below with reference to the accompanying drawings.
[0051] Such as figure 1 As shown, the pathogen molecule detection method based on nanopore sequencing provided by the embodiment of the present invention includes:
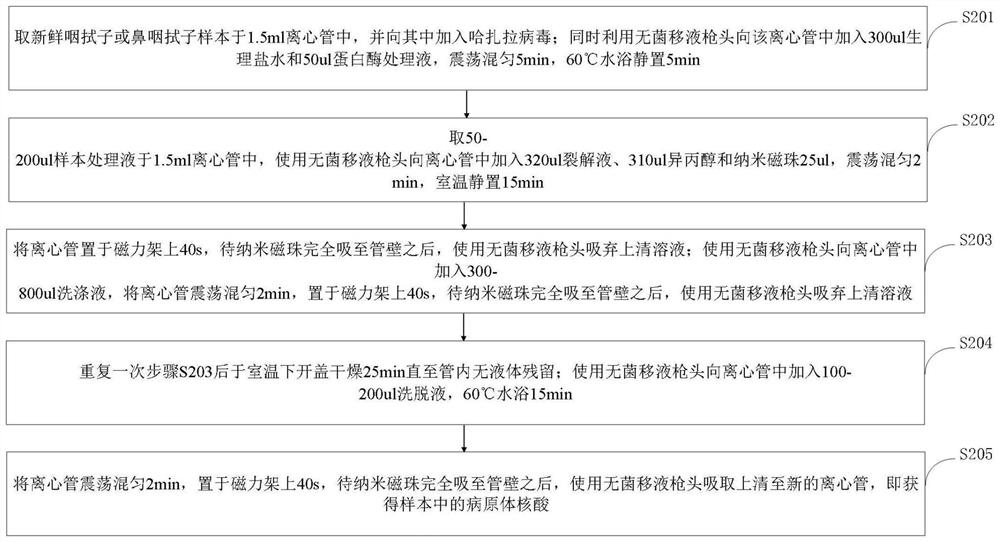
[0052] S101: Obtain a throat swab or nasopharyngeal swab sample; perform nucleic acid extraction and purification on the obtained throat swab or nasopharyngeal swab sample;
[0053] S102: Preparatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com