Recombinant escherichia coli for synthesizing 3-fucosyllactose and construction method thereof
A technology for recombining Escherichia coli and fucosyllactose, applied in the field of genetic engineering, can solve the problems of limited chemical properties of protective groups, expensive guanosine diphosphate fucose, unsuitable for large-scale food production, etc. Potential for industrial application, significant actual effect, good application prospect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Gene acquisition:
[0043] In the present embodiment, the gene manB (gene accession number GI: 946574) derived from Escherichia coli MG1655, the gene manC (gene accession number GI: 946580), the gene gmd (gene accession number GI: 946562), the gene fcl (gene accession number No. GI: 946563), gene lacY (gene accession number GI: 949083), and gene cafF (gene accession number GI: 187425317) derived from Akkermansia muciniphila ATCCBAA-835.
[0044] In this example, the E. coli genes manB, manC, gmd, fcl and lacY were successfully obtained by extracting the genomic DNA of E. coli MG1655; Gene cafF of Akkermansia mucinica ATCC BAA-835. The successful acquisition of the gene lays the foundation for the construction of the recombinant plasmid.
Embodiment 2
[0046] Preparation of recombinant plasmids
[0047] Use designed primers (upstream primer sequence as shown in SEQ ID NO.15, downstream primer sequence as shown in SEQ ID NO.16) the gene manC and gene manB derived from Escherichia coli MG1655 obtained in Example 1 are carried out PCR amplification , the amplified fragments were gel-cut and purified, and double-digested with NcoI and HindIII restriction endonucleases, and the digested fragments were ligated with the plasmid pCOLADuet-1, which was also double-digested with NcoI and HindIII, and Vector: The target fragments were mixed at a molar ratio of 1:3, added with T4 DNA ligase, and ligated for 5 hours at 22°C. The ligated product was transformed into E. coli DH5α competent cells, and screened on a kanamycin plate , to obtain the recombinant plasmid pCOLA-CB. Use designed primers (upstream primer sequence as shown in SEQ ID NO.17, downstream primer sequence as shown in SEQ ID NO.18) the gene gmd and gene fcl derived from E...
Embodiment 3
[0051] gene editing
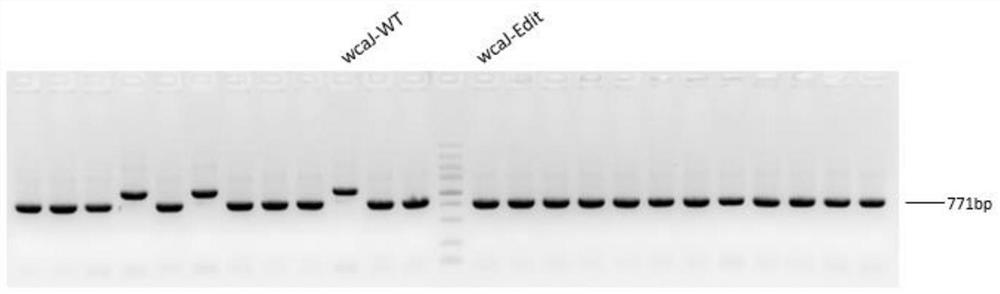
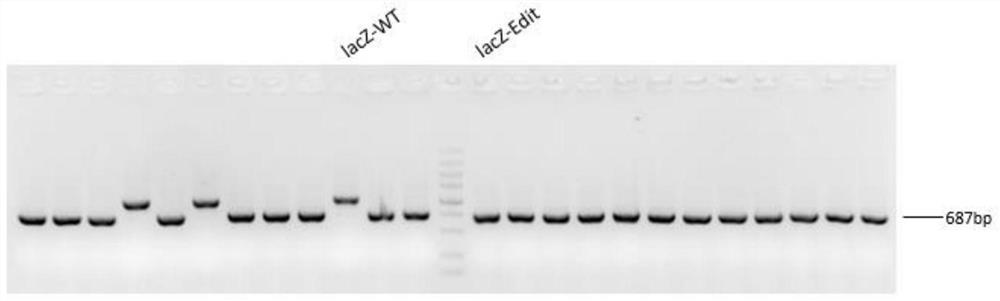
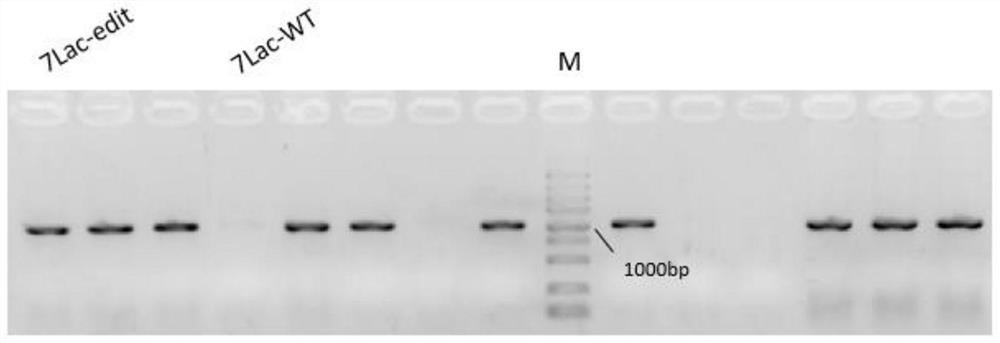
[0052] In this example, CRISPR gene editing technology was used to knock out the wcaJ and lacZ genes and to knock in the T7lac promoter. The steps of gene editing are described in detail below.
[0053] 1) The test showed that the original strain of Escherichia coli BL21 (DE3) was in normal growth state. The detection of target gene wcaJ and lacZ and T7lac promoter knock-in positions and their upstream and downstream sequences showed that the size of PCR amplified bands was in line with expectations, and the sequencing results were consistent with NCBI Sequence is consistent;
[0054] 2) According to the insertion position of the target gene sequence and the surrounding sequence characteristics, design and prepare gRNA, clone the gRNA and Donor sequence into the gene editing vector Donor plasmid, and verify by sequencing to ensure that the gRNA and Donor sequence in the constructed vector are consistent with the target sequence consistent;
[0055] 3) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com