Oudemansiella radicata SSR molecular marker primer group and application thereof
A technology of molecular markers and long-rooted mushrooms, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problems of long time, morphological changes of edible fungus fruiting bodies, etc., and achieve high accuracy and expansion Increase the effect of clear and specific bands
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Example 1: SSR locus analysis, primer screening and cluster analysis of Rhizome lanceolata
[0080] 1. SSR locus analysis of Rhizoma lanceolata
[0081] The whole genome DNA of Rhizome lanceolata samples was extracted by conventional CTAB method, and the whole genome was sequenced by Illumina HiSeq 2000 sequencer, and the obtained genome size was 57.3Mb. The SSRHunter software was used to search for SSR sequences on the Rhizoma lanceolata genome, and the screening conditions were set as follows: the number of repeated bases was 2-6, and the number of repeats was greater than or equal to 5 times, and a total of 1144 SSR sequences were obtained.
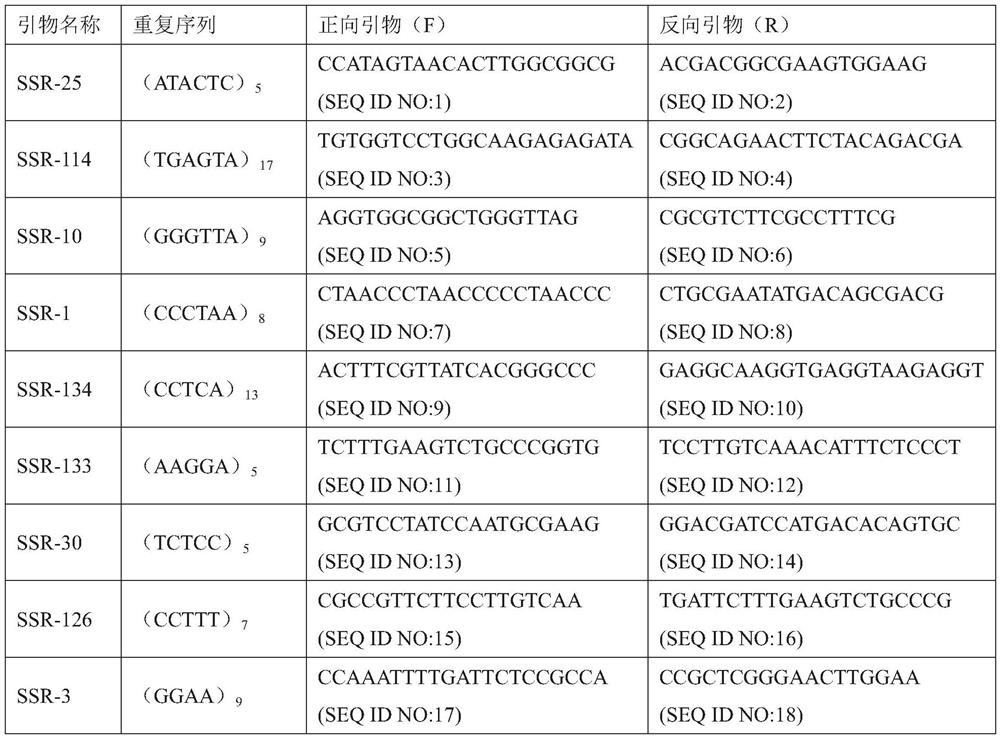
[0082] 2. Primer design
[0083] Primers were designed within the range of 200 bp upstream and downstream of the detected SSR site, and the primer design conditions were as follows: the length of the primer was between 18-25 bp, and the annealing temperature was 60±3°C. A total of 20 pairs of primers were designed, and the pri...
Embodiment 2
[0097] Example 2: Application of SSR-labeled primers of Rhizoma lanceolata in strain identification
[0098] 1. The genomic DNA of the strain was extracted by conventional CTAB method. The tested strains were the mycelium of Rhizoma lanceolata, Lentinus edodes, Pleurotus eryngii, Agaricus bisporus, Ganoderma lucidum, Fungus, and Shiji mushroom strains.
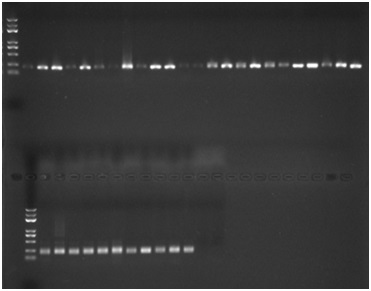
[0099] 2. The extracted genomic DNA is used as a template, amplified with SSR-133 primers, and detected by electrophoresis.
[0100] PCR reaction system: Genomic DNA as template 0.5 microliters, SSR-133F upstream primer 2 microliters, SSR-133R downstream primer 2 microliters, 2×pfu mix 12.5 microliters, ddH 2 O to make up to 25 μl.
[0101] PCR reaction program: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 1 min, degradation at 60°C for 1 min, extension at 72°C for 1 min, and repeat the process 35 times; extension at 72°C for 10 min; incubation at 4°C.
[0102] PCR product detection: PCR amplification produc...
Embodiment 3
[0104] Embodiment 3: SSR identification of different parts of the same bacterial strain
[0105] 1. Extract the genomic DNA of the stipe, mushroom root and mushroom part of the fruiting body of the mushroom;
[0106] 2. Using the extracted genomic DNA as a template, use SSR primers to amplify and detect by electrophoresis;
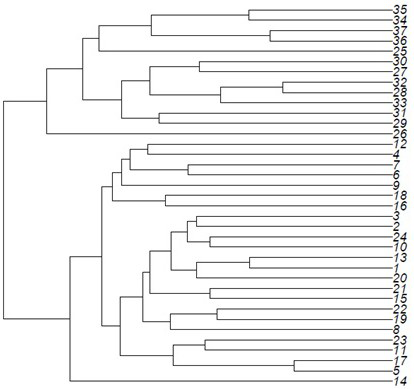
[0107] 3. The electrophoretic bands are consistent with the electrophoretic bands amplified by mycelium, and the layer clustering analysis shows that the similarity coefficient is 100%, and they are completely clustered together, which verifies the effectiveness of the method.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com