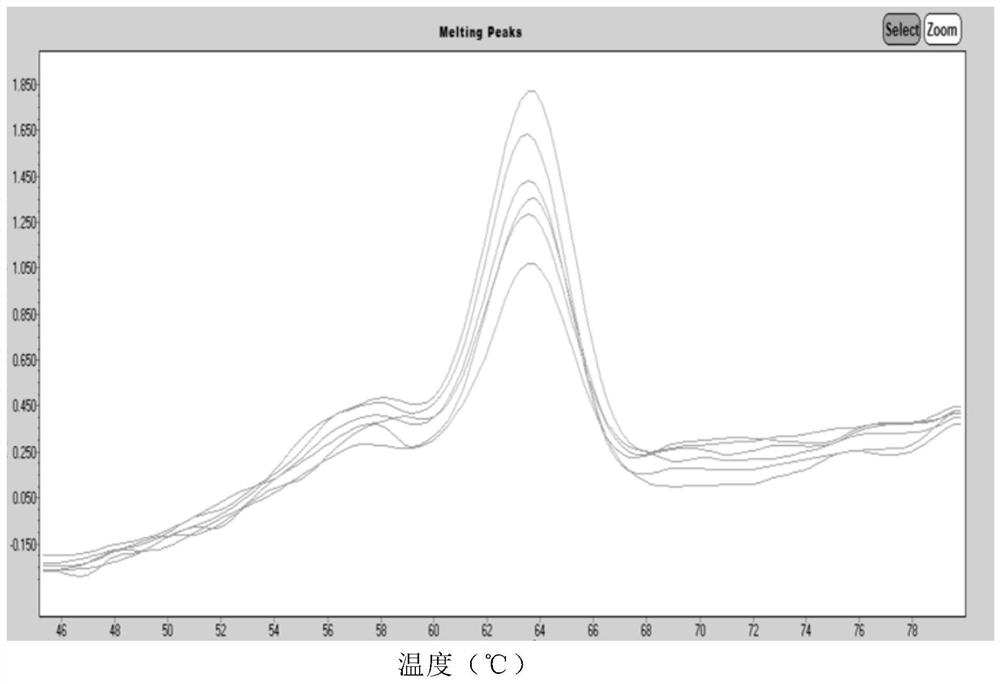
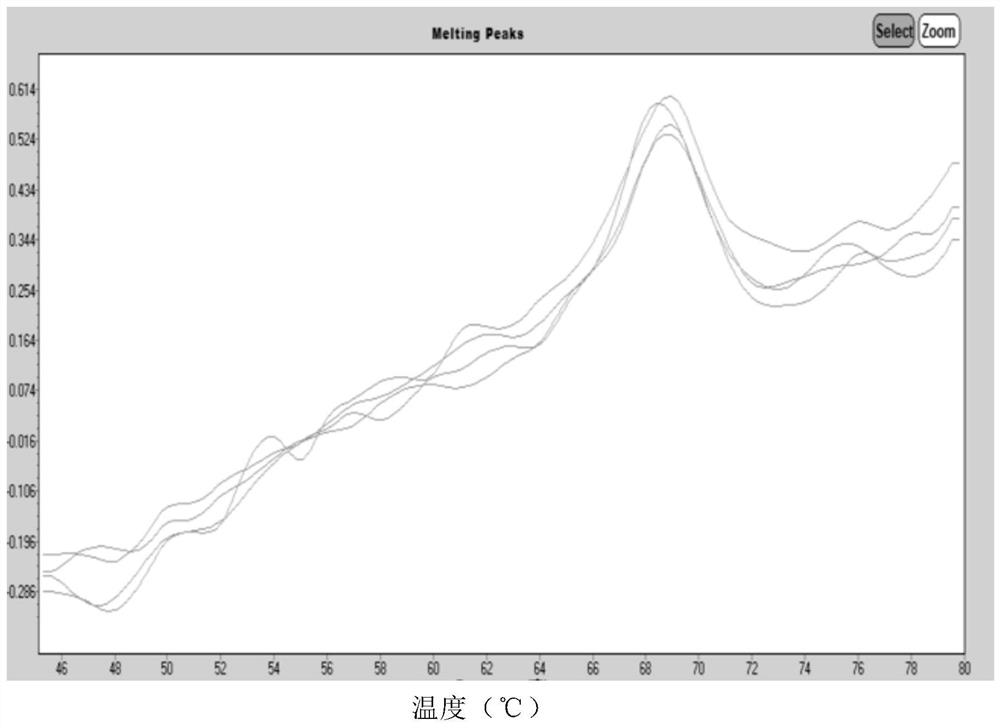
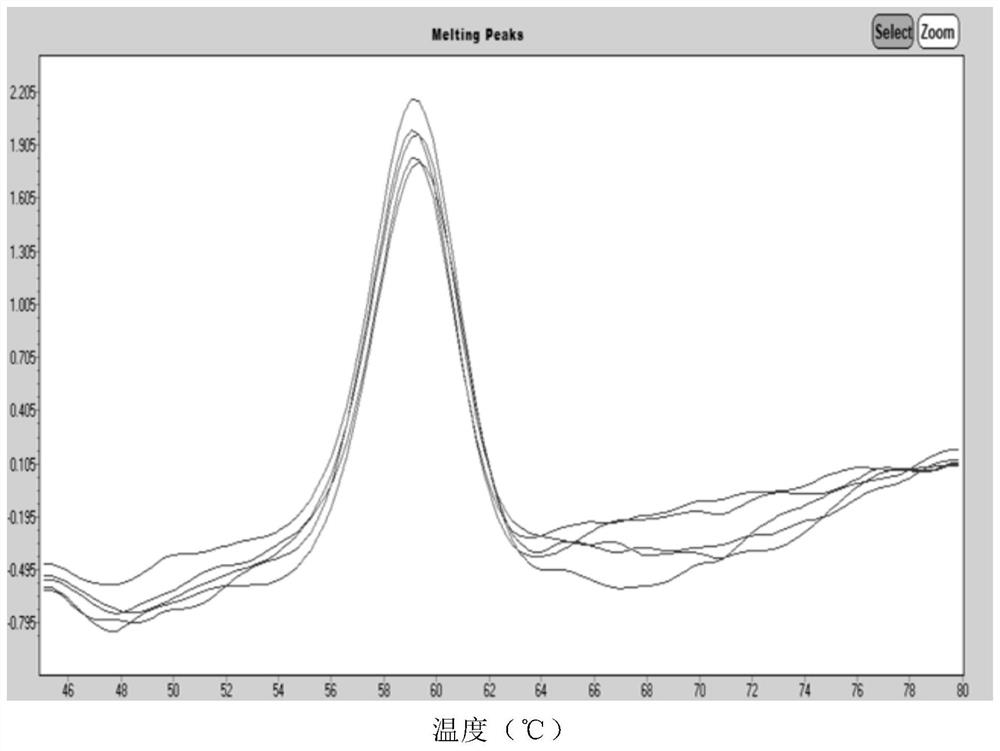
Human coronavirus nucleic acid multiplex detection kit
A coronavirus, multiplex detection technology, used in recombinant DNA technology, microbial assay/test, resistance to vector-borne diseases, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] The detection method of the human seven-fold coronavirus nucleic acid detection kit is as follows:
[0098] Samples: Various respiratory samples such as swabs, sputum, alveolar lavage fluid
[0099] Storage conditions: room temperature (18-30°C), no more than 48 hours; refrigerated temperature (2-8°C), no more than 72 hours; below -20°C, long-term storage is possible.
[0100] Steps:
[0101] 1. Extraction of sample nucleic acid.
[0102] Add 5uL internal quality control (IC) to 200uL sample, extract nucleic acid and finally elute with 100uL volume. Note that the negative control (NC) provided in the kit should be added to the internal quality control (IC) like other test samples before nucleic acid extraction. The positive control (PC) in the kit does not require nucleic acid extraction.
[0103] 2. rRT-PCR steps
[0104] 2.1 Preparation of rRT-PCR reaction solution.
[0105] Dissolve CoV detection buffer and CoV enzyme mixture at room temperature, flick to mix a...
Embodiment 2
[0125] Embodiment 2 Specificity and Sensitivity Determination
[0126] specificity test
[0127] Influenza A virus, influenza B virus, respiratory syncytial virus, adenovirus, Chlamydia pneumoniae, Mycoplasma pneumoniae, Streptococcus pneumoniae, Legionella pneumophila, Haemophilus influenzae as positive templates, cytomegalovirus, simple Herpesvirus type 1, human coronavirus NL63, human bocavirus, Epstein-Barr virus, parainfluenza virus type 1, metapneumovirus, enterovirus / rhinovirus, Mycobacterium tuberculosis, Escherichia coli, Pseudomonas aeruginosa nucleic acid is The negative template was tested by multiple detection kits for respiratory pathogens. The results showed that the detection reagents could accurately detect influenza A virus, influenza B virus, respiratory syncytial virus, adenovirus, Chlamydia pneumoniae, Mycoplasma pneumoniae, Streptococcus pneumoniae, Nucleic acid of Legionella pneumoniae and Haemophilus influenzae, and there was no false detection or miss...
Embodiment 3
[0130] Embodiment 3 clinical sample detection
[0131] Using the multiple detection system for respiratory pathogens established by the present invention, 60 cases of positive patient nucleic acid samples and 20 cases of negative samples were detected, and a commercially available single-plex PCR detection kit was used for simultaneous detection, and the positive samples were sequenced and verified .
[0132] The results show that among the 60 positive samples, commercially available single-plex fluorescent quantitative PCR and multiple fluorescent PCR methods of the present invention all detect 60 positive samples, and the positive detection rate is 100%, and 20 negative sample tests are all negative. The method consistency was high, and the results were statistically significant.
[0133] Specific to different types, the results showed 6 cases of 2019-nCoV virus, 1 case of MERS-CoV virus, 23 cases of OC43 virus, 11 cases of 229E virus, 13 cases of NL63 virus, and 6 cases of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com