Affinity peptide M1 of novel corona virus main protease and application thereof
A coronavirus, main protease technology, applied in the field of biomedicine, can solve problems such as false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084]Example 1 The virus main protease M of SARS-CoV-2 pro Preparation of the affinity peptide
[0085] 1. Experimental materials
[0086] Ph.D.-12 Phage Display Peptide Library Kit was purchased from New England Biolabs, Inc. Interaction kinetics analysis and affinity determinations used ForteBio Octet instruments. The experimental reagents and experimental operation methods were carried out in accordance with the instructions of the product.
[0087] 2. Experimental method
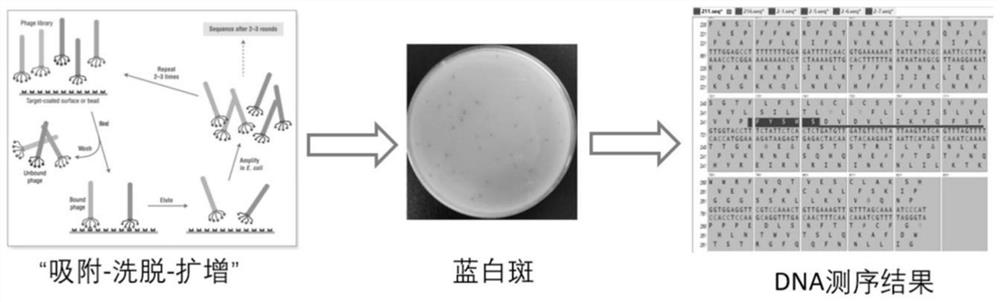
[0088] Obtaining the main virus protease M of SARS-CoV-2 by phage display technology pro Affinity peptides, using M13 phage, pIII display system peptide library, the library includes 10 9 different polypeptide sequences. Repeat the process of "adsorption-elution-amplification" for 3-5 rounds to obtain phages that can bind to the target protein. Extract the DNA of the phage, complete the sequencing, analyze and obtain the polypeptide sequence that can bind to the target protein, and further synthe...
Embodiment 2
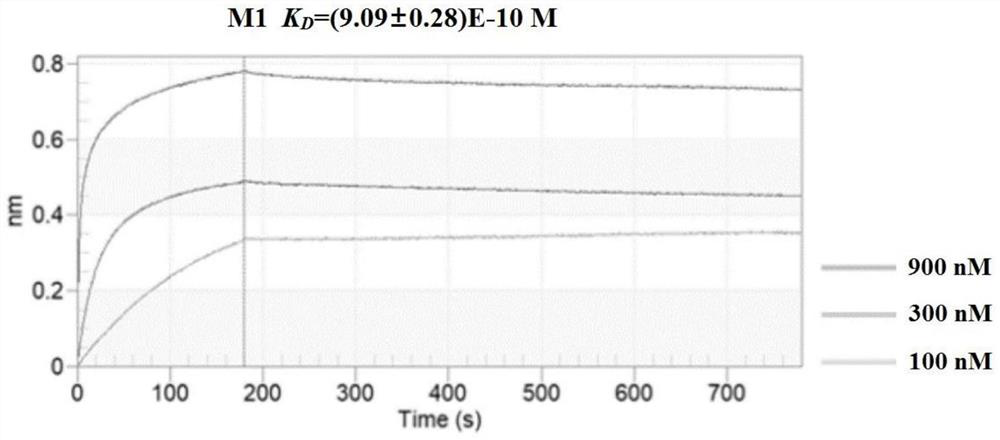
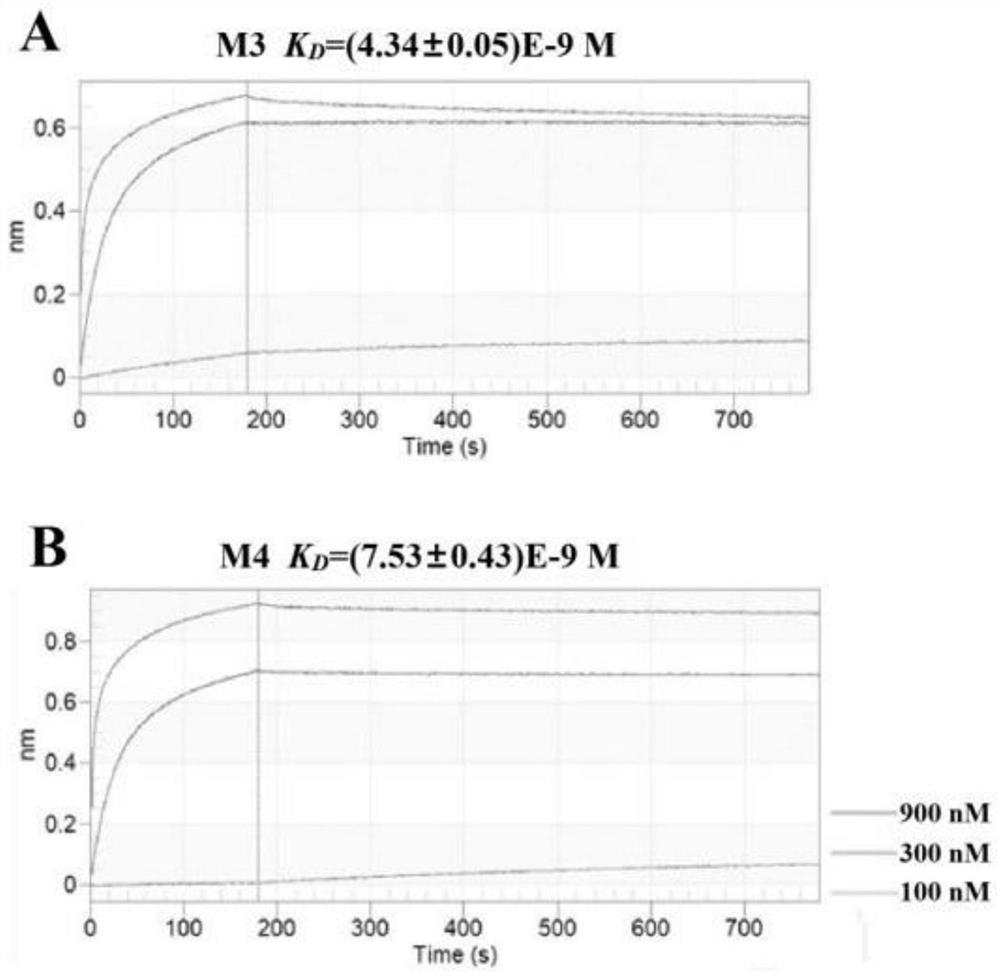
[0149] Example 2 Detection of Affinity and Interaction Kinetics between Affinity Peptide and Target Protein
[0150] 1. Experimental method
[0151] Detection of affinity peptides M1, M3, M4 and target protein M by Bio-layer Interferometry (BLI) pro The affinity of the interaction between them, and the kinetic process of the binding and dissociation between the polypeptide and the target protein. Modify the M1 polypeptide at the C-terminal with Biotin (Biotin-GG-GDAH TRPP WSAW-NH 2 ), immobilized on the SA-modified sensor, the treatment of M3 and M4 in the control group was the same as that of M1, and it was plotted against different concentrations of M pro The binding and dissociation kinetic curves during the interaction, the dissociation equilibrium constant K of their interaction is calculated from the binding and dissociation kinetic curves D , the detailed experimental steps are as follows:
[0152] (1) Immerse the biosensor surface-modified with streptavidin (Strept...
Embodiment 3
[0163] Example 3 Verification of the responsiveness of the affinity peptide M1 to the target protein
[0164] 1. Experimental method
[0165] A cysteine is introduced at the C-terminus of the affinity peptide, so that the peptide is bound to the surface of gold nanoparticles through Au-S bonds. Add a series of concentration gradient target proteins to the system, and detect the SPR peak position of the gold nanoparticles when the protein binds to the affinity peptide on the surface of the gold nanoparticles. The detailed experimental steps are as follows:
[0166] (1) Preparation of gold nanoparticles
[0167] Add 150mL sodium citrate solution (2.2mM) into a 250mL round bottom flask, place in a water bath at 90°C, stir magnetically at 400rpm, add 1mL chloroauric acid solution (25mM), react for 30min, and then add 1mL 60mM citric acid Sodium solution and 1mL 25mM chloroauric acid solution, after reacting for 30min, add 1mL 60mM sodium citrate solution and 1mL 25mM chloroau...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com