Two-step PCR technology
A technical and specific technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of time-consuming and other problems, and achieve the effect of short time-consuming, wide adaptability and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
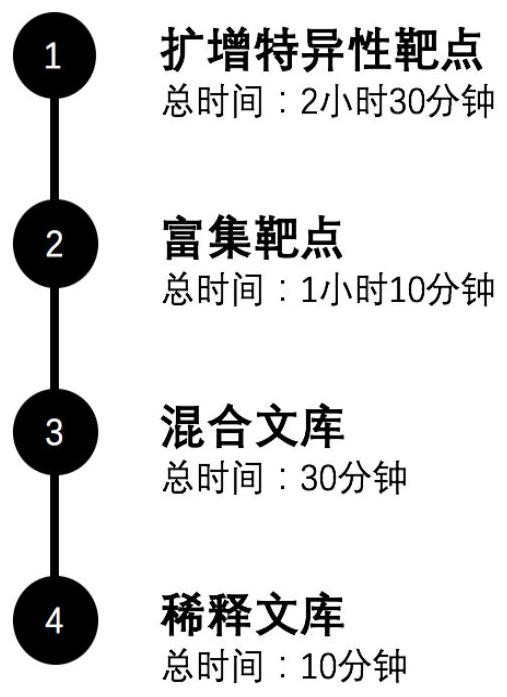
[0027] In an embodiment of the present invention, a two-step PCR technique comprises the following steps:
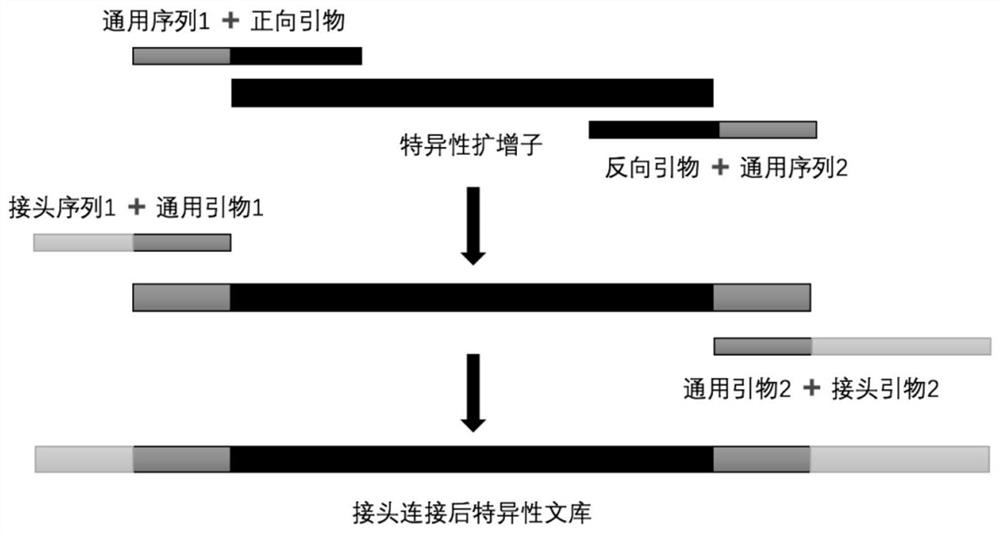
[0028] The first step is to amplify the specific target: the DNA sample and the specific primer pair mixture are mixed in this part, so that the universal sequences of the forward and reverse primers are respectively connected to the two ends of the specific amplicon determined by the specific primers. On the other hand, this part of the reaction is also called the first step of PCR.
[0029] The second step is to enrich the target: in the enrichment of the target, which is the second step of PCR, we connected the linker after the universal sequence connected in the first step.
[0030] The third step is to mix the library: this part uses the fluorescence quantitative method to quantify the enrichment products, and records the sample information and concentration information.
[0031] The fourth step is to dilute the library: use the fluorescent quantitative method to q...
Embodiment 2
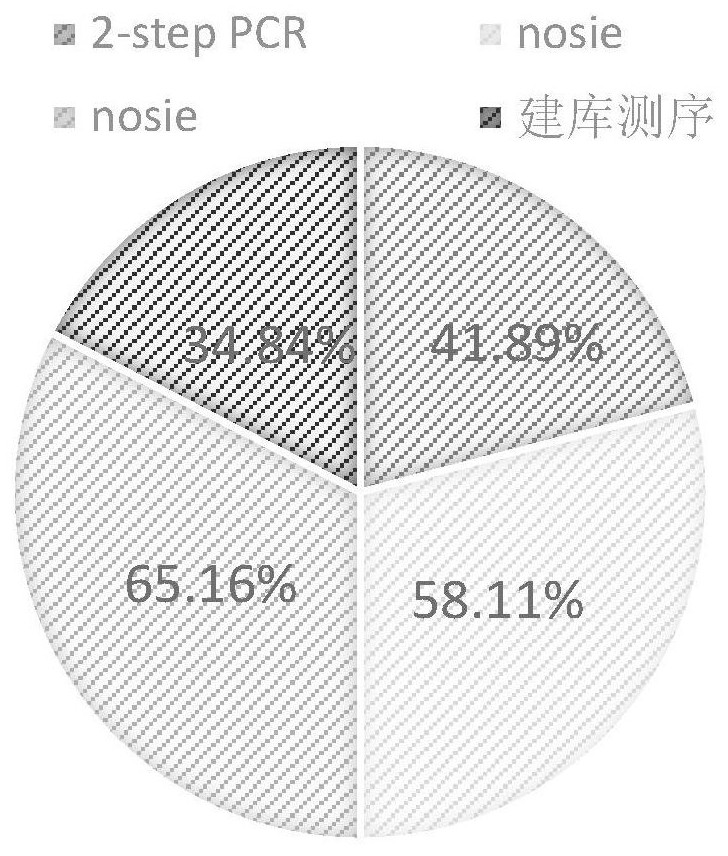
[0037] image 3 It is a graph showing the proportion of data utilization under different methods, where noise represents sequencing noise.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com