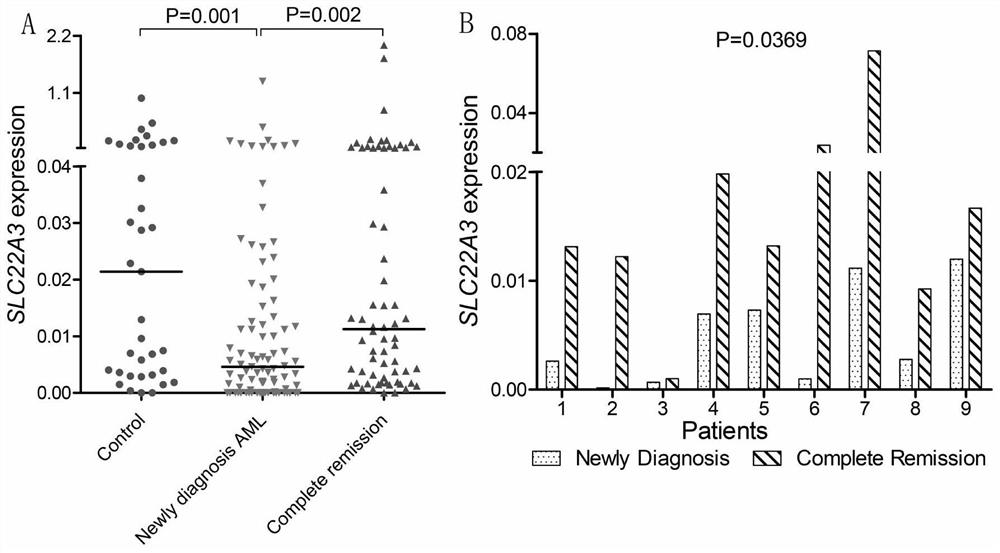
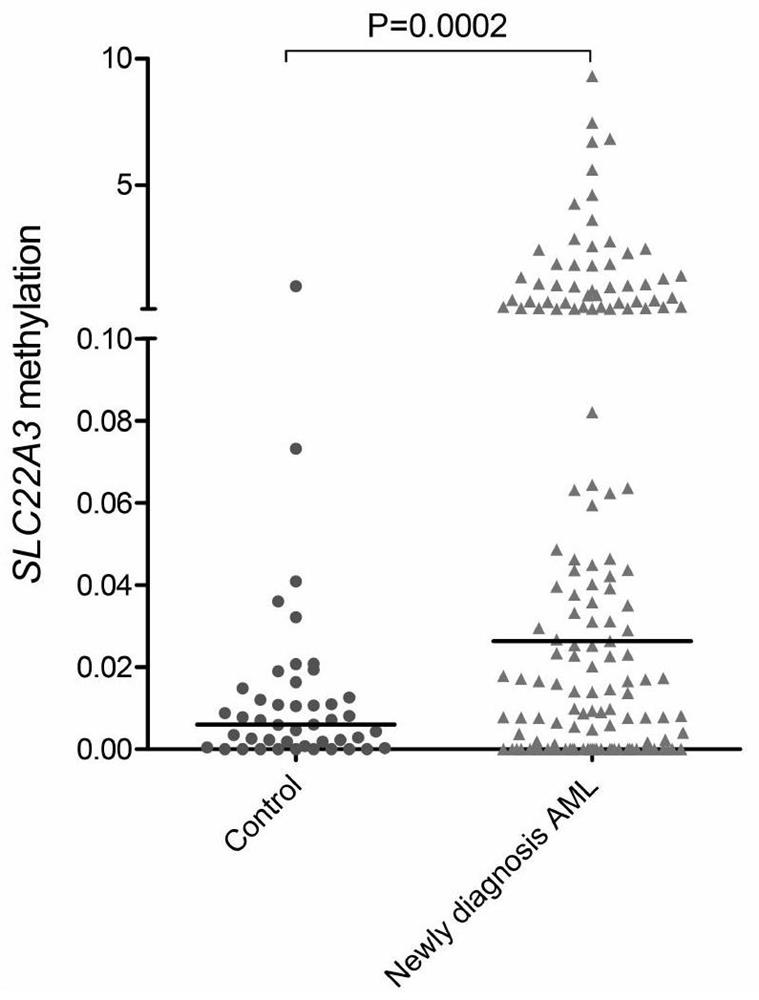
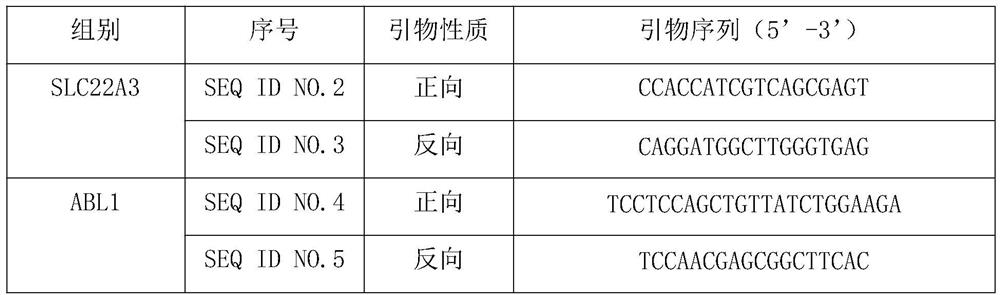
Quantitative detection method and application of molecular marker SLC22A3
A technology for detecting substances and drugs, which is applied in the field of molecular biology and can solve problems such as meanings that need to be discovered urgently
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Establishment of a method for detecting the expression level of SLC22A3 based on quantitative PCR
[0032] (1) Total RNA extraction:
[0033] ① Separation of bone marrow mononuclear cells: Add 5ml of lymphocyte separation medium and red blood cell lysate to the obtained bone marrow specimen successively, mix upside down, centrifuge at 1000×rpm for 5min, and discard the supernatant;
[0034] ② Add 1ml TRIZOL to the above precipitate, and fully pipette and mix until the water sample;
[0035] ③Add 200μl chloroform, vortex fully shake for 30sec, let stand at room temperature for 5min, and centrifuge at 12000×rpm for 10min;
[0036] ④Add the supernatant to a clean 1.5ml EP tube, add 600μl isopropanol at the same time, close the lid tightly, mix up and down 50 times, let stand at room temperature for 10min, centrifuge at 12000×rpm for 10min, carefully discard the supernatant, and transfer to the EP tube The bottom white precipitate is RNA;
[0037] ⑤ Add 1ml of...
Embodiment 2
[0052] Example 2: Design and synthesis of primers for specifically amplifying the CpG island of the SLC22A3 gene promoter region
[0053] Aiming at the CpG island information in the promoter region of SLC22A3 gene, two sets of primers were designed for methylated and unmethylated sequences. These two pairs of primers can be used to specifically amplify the methylated and unmethylated sequences of the CpG island in the promoter region of SLC22A3 respectively. Methylation sequence, followed by quantitative analysis of methylation, the specific design steps are as follows:
[0054] The SLC22A3 promoter CpG island sequence was found and exported at UCSC (http: / / genome.ucsc.edu / ):
[0055] Path: UCSC homepage→Tools→gene sorter→Genome(Human)→search(SLC22A3)→go→select SLC22A3→Genomic Sequence→select the 5’UTR of 2000bp upstream and 60bp downstream of the promoter→export the obtained sequence.
[0056] Use the Methyl primer 1.0 software to design the required MSP primers:
[0057] I...
Embodiment 3
[0060] Example 3: Establishment of a method for detecting the methylation status of the SLC22A3 gene promoter GpG island based on quantitative PCR
[0061] The detection method of the present invention is based on the principle of methylation-specific PCR, and methylated and non-methylated DNA sequences can be distinguished through quantitative PCR amplification containing methylated and unmethylated specific primers respectively . The method comprises the steps of:
[0062] (1) DNA template preparation
[0063] Take 5-10 ml of heparin-anticoagulated bone marrow specimens from AML and normal controls, use Ficoll solution to separate mononuclear cells, and use a genomic DNA extraction kit (purchased from Gentra Company) to extract genomic DNA in mononuclear cells. The specific steps are as follows:
[0064] ① Add 1ml TRIZOL (lysate) to the cells and mix well until the water sample;
[0065] ② Add 200 μl of chloroform (chloroform), close the lid tightly, shake and mix well on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com