Recombinant strain for modifying corynebacterium glutamicum promoter, construction method of recombinant strain and application of recombinant strain in production of L-amino acid
A technology of Corynebacterium glutamicum and promoter, applied in the fields of genetic engineering and microorganisms, can solve problems such as product accumulation, and achieve the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Embodiment 1 constructs the pEC-H10-mCherry plasmid that strong promoter H10 promotes the expression of mCherry in Corynebacterium glutamicum
[0052]The strongest promoter H10 in Corynebacterium glutamicum reported by Wei et al. (SEQ ID NO:11, GCTAACCCTTACCGGTCGGCTCTAAGCCGGCGGCGTATGGTAAGCTCTGTTATGTATA GTCCGAGCACGGCGAAAGGATACTC) as a template, and the gene sequence of mCherry protein and the sequence of expression vector pEC-XK99E in Corynebacterium glutamicum, designed and synthesized primers for the construction of pEC-H10-mCherry vector. The primers were designed as follows (synthesized by Guangzhou Jinweizhi Co.):
[0053]
[0054] Construction method: using pEC-XK99E as a template, using primers 1 (SEQ ID NO:1) and 2 (SEQ ID NO:2) to amplify the backbone region (6743bp) of the pEC plasmid, and using primer 3 (SEQ ID NO:3) and 4 (SEQ ID NO:4) amplify the fragment (387bp) containing the upstream region of the H10 promoter; with the plasmid pBblactam containing th...
Embodiment 2
[0055] Example 2 Constructing a Saturation Mutation Library of Strong Promoter H10
[0056] Using the pEC-H10-mCherry plasmid as a template, the non-conserved region (NNAGGANNNNN) of the RBS binding site in the H10 promoter region and the 5 base regions upstream were designed and synthesized for the saturation mutation library primers. The primers were designed as follows (Guangzhou Golden Synthesized by Weizhi Company):
[0057]
[0058] Construction method: using the pEC-H10-mCherry plasmid as a template, primers 3 and 7 (SEQ ID NO: 7) amplified the fragment containing the saturation mutation in the upstream region of the H10 promoter, and primers 8 (SEQ ID NO: 8) and 6 amplified To increase the saturation mutation fragment containing the RBS binding site, the PCR reaction system is: 0.5 μl template DNA, 0.25 μl each primer (100 pmol), 25 μl pfu (Novazyme 2×Phanta Master mix), dH 2 O 24 μl, total volume 50 μl. The PCR amplification was carried out as follows: 95°C pre-d...
Embodiment 3
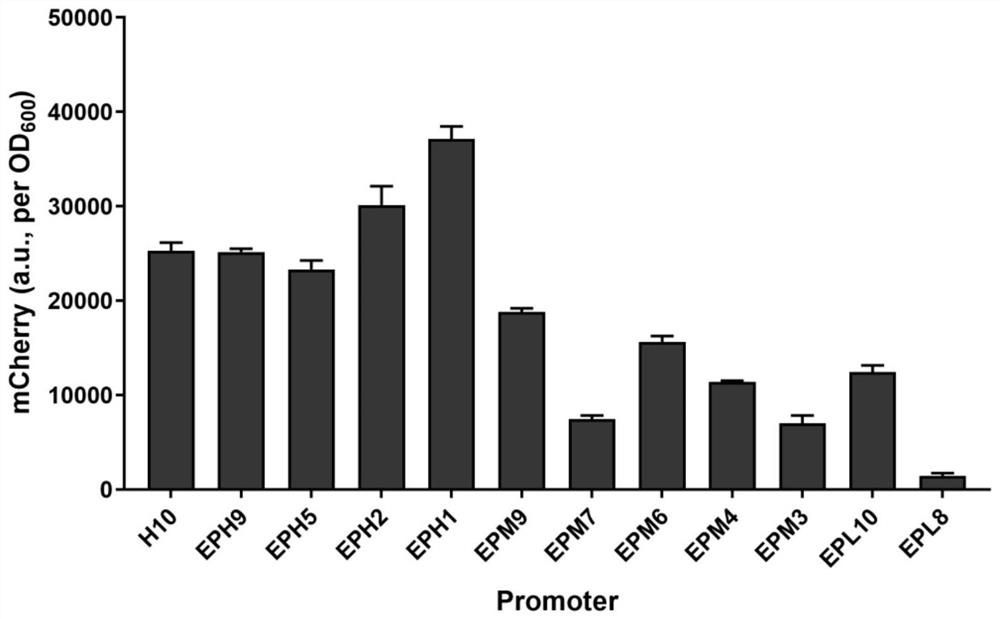
[0059] Example 3 Screening for a more active promoter from the H10 promoter mutation library
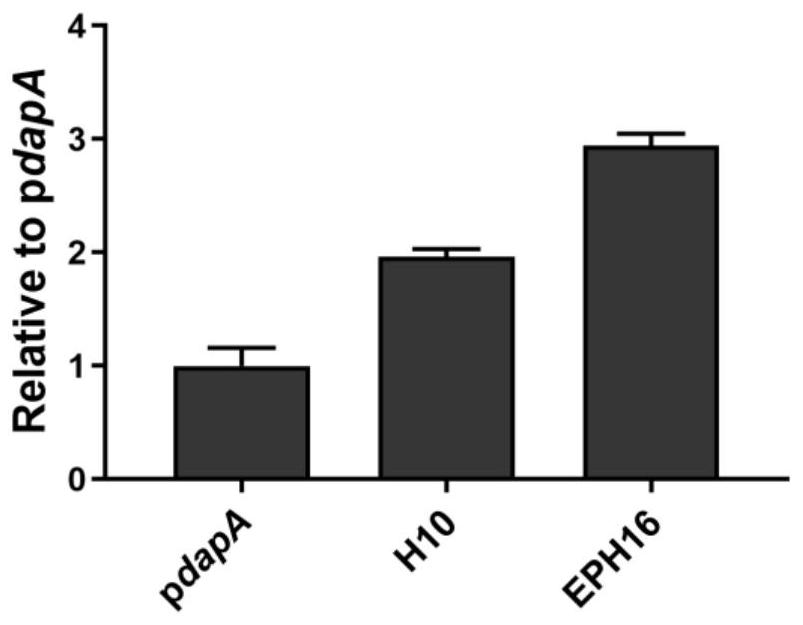
[0060] From the H10 promoter mutation library in Corynebacterium glutamicum obtained in Example 2, randomly select clones to contain 96 deep-well plates of 900 μl LBHIS medium, and select 5 96 deep-well plates altogether, wherein each 96 Each deep well plate contains 3 unmutated H10 promoter strains. After cultured at 30°C and 800rpm for 24 hours, use a microplate reader to detect the fluorescence value of mCherry, and screen out the promoter with stronger activity than the H10 promoter according to the fluorescence intensity. , named the stronger promoter that was screened out as EPH16, and sequenced it. The sequence was (SEQ ID NO: 9): GCTCAgctttTACCGGTCGGCTCTAAGCCGGCGGCGTATGGTAAGCTCTGTTATGTATAGTCCGAGCACGGCGAAAGGATGAAT.
[0061] Each promoter of H10, EPH16 and pdapA was used as a reporter protein with mCherry, and the vector was constructed based on the pEC plasmid; the competent s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com