Method for detecting novel coronavirus variants and subtypes
A technology for coronaviruses and mutant strains, applied in the fields of probes, detection systems, and specific crRNA, can solve the problems of high technical and professional knowledge requirements for operators, high configuration requirements, and long time-consuming, etc., to achieve shortened detection time, high sensitivity, The effect of easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] A specific crRNA used to identify the nucleic acid of different coronavirus SARS-CoV-2 variants, based on CRISPR-Cas12a technology detection,
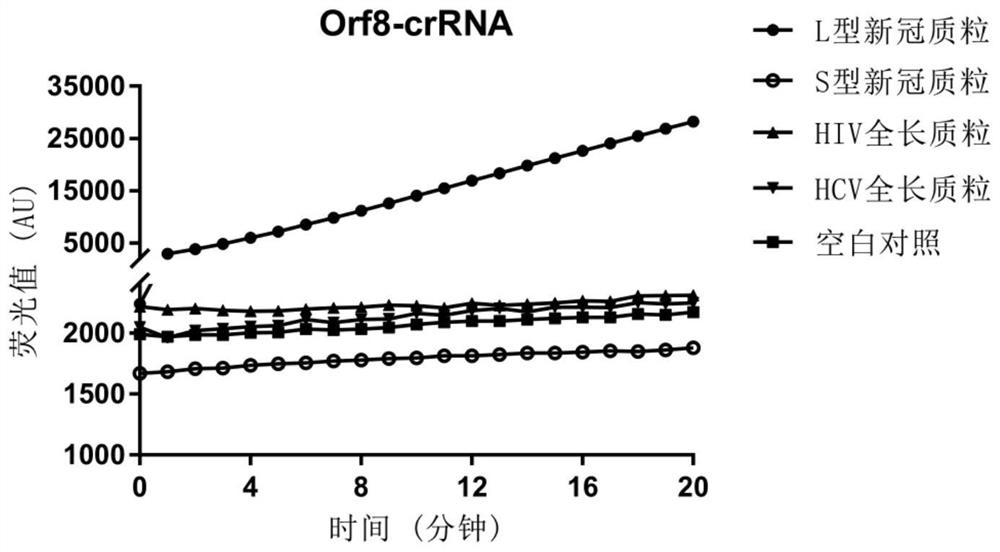
[0067] The specific crRNA used to identify the L subtype and the S subtype is Orf8-crRNA, and the nucleotide sequence is 5'-UAAUUUCUACUAAGUGUAGACCUUUUACAAUUAAUUGCCA-3' (SEQ ID NO 1).
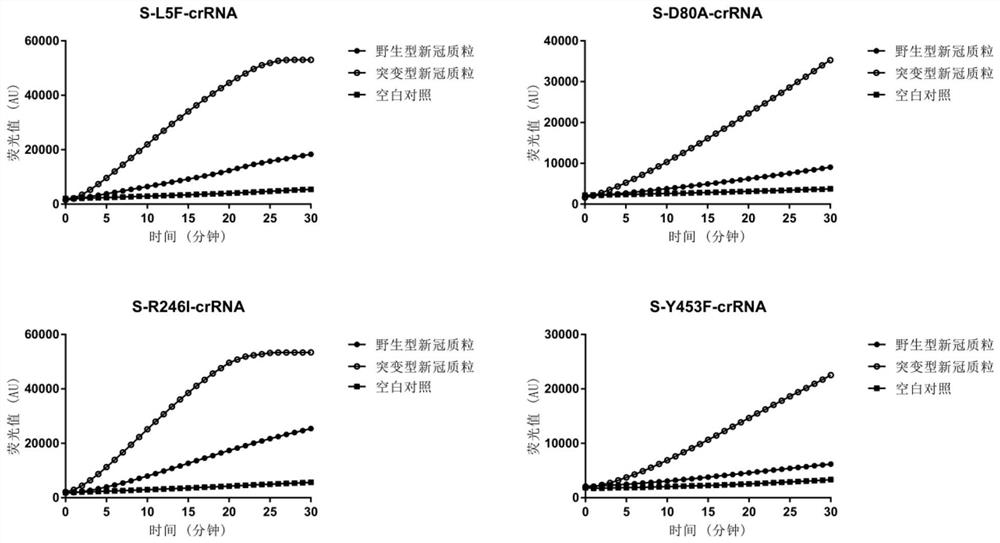
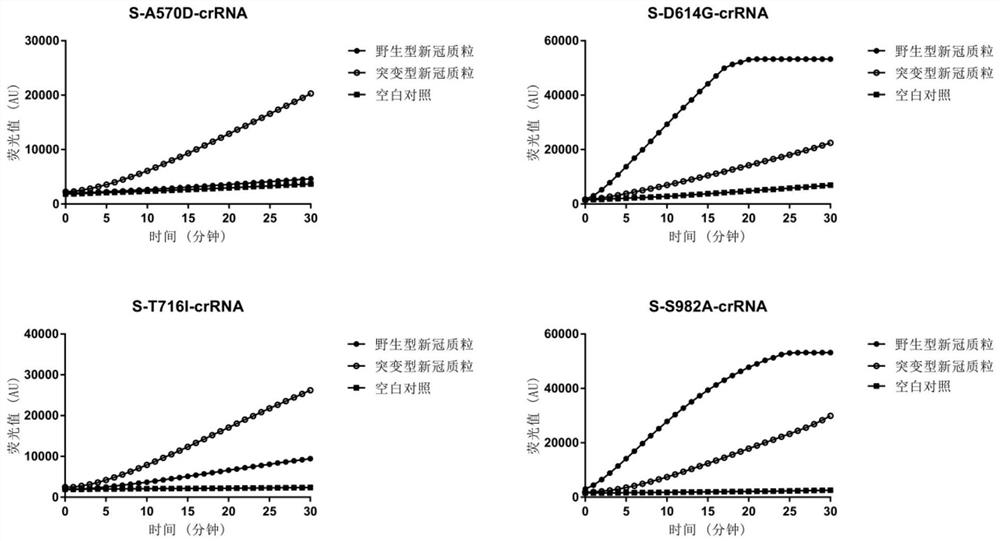
[0068] The specific crRNAs used to identify the mutation sites L5F, D80A, D215G, R246I, Y453F, N501Y, A570D, D614G, A701V, T716I, S982A, P1263L, K417N, L452R, and E484Q of the S protein gene of SARS-CoV-2 are S-L5F-crRNA, S-D80A-crRNA, S-D215G-crRNA, S-R246I-crRNA, S-Y453F-crRNA, S-N501Y-crRNA, S-A570D-crRNA, S-D614G-crRNA, S- A701V-crRNA, S-T716I-crRNA, S-S982A-crRNA, S-P1263L-crRNA, S-K417N-crRNA, S-L452R-crRNA, S-E484Q-crRNA, the nucleotide sequences are respectively:
[0069] S-L5F-crRNA: 5'-UAAUUUUCUACUAAGUGUAGAUUUUAUUGCCACUAGUCUCU-3' (SEQ ID NO2);
[0070] S-D80A-crRNA: 5'-UAAUUUCUACUAAGUGUAGACUAACCCUGUCCUACCAUUU-3' (SEQ ID NO.3);
[0071] ...
Embodiment 2
[0107] A CRISPR-Cas12a detection system for detecting variants of SARS-CoV-2, including the specific crRNA in Example 1, probes, nuclease-free water and buffer buffer2.1.
[0108] The detection system is detected through the following steps:
[0109](1) Using the detection system, the detection system includes 0.4 μL of crRNA with a concentration of 15 μM, 1 μL of Cas12a with a concentration of 10 μM, 1 μL of a reporter probe with a concentration of 10 μM, 1× buffer buffer 2.1 27.6 μL, and placed at a constant temperature of 37°C After incubation in the fluorescence detector for 10 minutes, add SARS-CoV-2 genomic DNA as a template for CRISPR-Cas12a detection, the detection time is 30 minutes, and the detection result is obtained;
[0110] (2) Analyze the fluorescent signal to determine whether the sample contains SARS-CoV-2 variant virus nucleic acid. Fluorescent signals are detected and judged as positive, indicating that there is nucleic acid of the corresponding variant st...
Embodiment 3
[0117] The CRISPR-Cas12a detection method of the present invention is used to identify SARS-CoV-2-Orf8-L type / S type.
[0118] This example is used to detect the CRISPR-Cas12a detection system of SARS-CoV-2 mutant strains, which contains SARS-CoV-2 specific crRNA, universal probe, nuclease-free water, and buffer 2.1. Buffer buffer 2.1 (New England BioLabs, Ipswich, M0653T), nuclease-free water (Aikerui Bio, AG11012). crRNA (synthesized by Qingke Biotechnology Co., Ltd.) was used at a concentration of 15 μM, Cas12a (New England BioLabs, Ipswich, M0653T) was used at a concentration of 10 μM, and the probe (synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.) was used at a concentration of 10 μM. 1× buffer buffer2.1.
[0119] crRNA and probe sequences are as follows:
[0120] Orf8-crRNA:5'-UAAUUUCUACUAAGUGUAGACCUUUUACAAUUAAUUGCCA-3'(SEQ ID NO.1)
[0121] Probe sequence: 5'-TTATT-3' (SEQ ID NO.14)
[0122] Entrust Sangon Bioengineering (Shanghai) Co., Ltd. to synthesize ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com