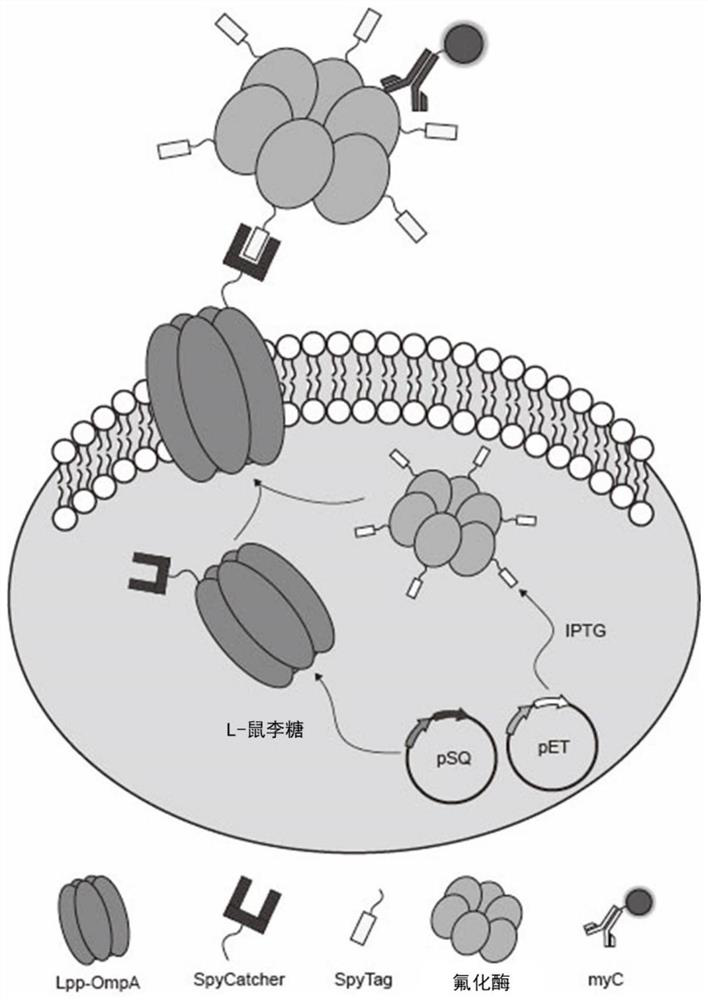
Method for surface display of fluorinase based on self-assembly
A surface display and self-assembly technology, applied in the field of genetics and bioengineering, can solve the problems of loss of enzyme catalytic activity, limitation of protein size and complexity, protein inactivation, etc., to avoid purification steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1: Construction of SC-ST surface display system
[0037] Using the plasmid co-expression system, two plasmids with different replicons and resistances were selected to express the target protein. For the anchoring protein Lpp-ompA, a strictly controlled rhaB rhamnose-inducible promoter was selected, and RBScalculator was used to design and modify the RBS on the expression vector pSQ036 (the RBS sequence is shown in SEQ ID NO.1). For the reported genes flA, SpyCatcher, SpyTag, and Lpp-OmpA, gene synthesis was carried out (see Table 1 for the nucleotide sequence), and they were respectively connected to T vectors for preservation. Primers were designed to amplify the target gene from the T vector, and the PCR product was purified after verification by DNA agarose gel to obtain the cloned target gene. Using the Gibson assembly method, TriggerFactor (gene number WP_001198386.1), Lpp-OmpA, and SpyCatcher genes were connected to the expression vector pSQ036 to con...
Embodiment 2
[0042] Example 2: Heterologous Expression of Fluorase in E. coli
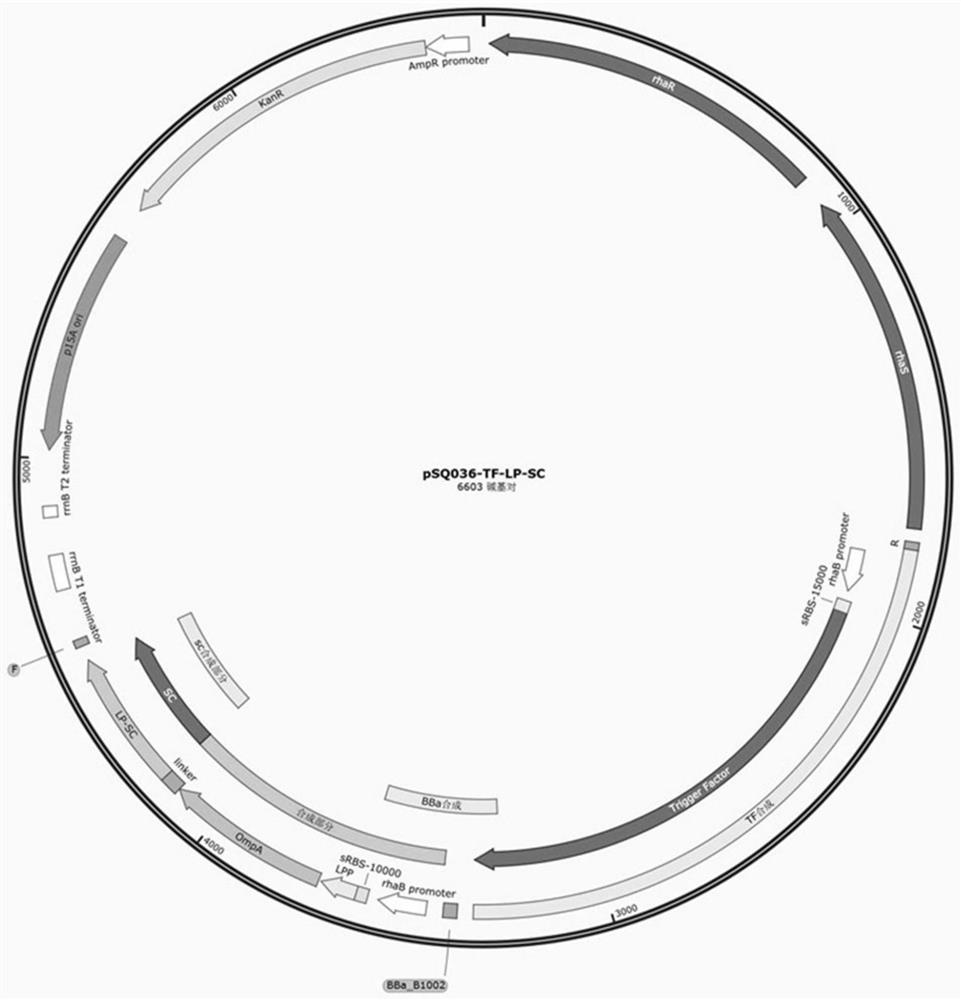
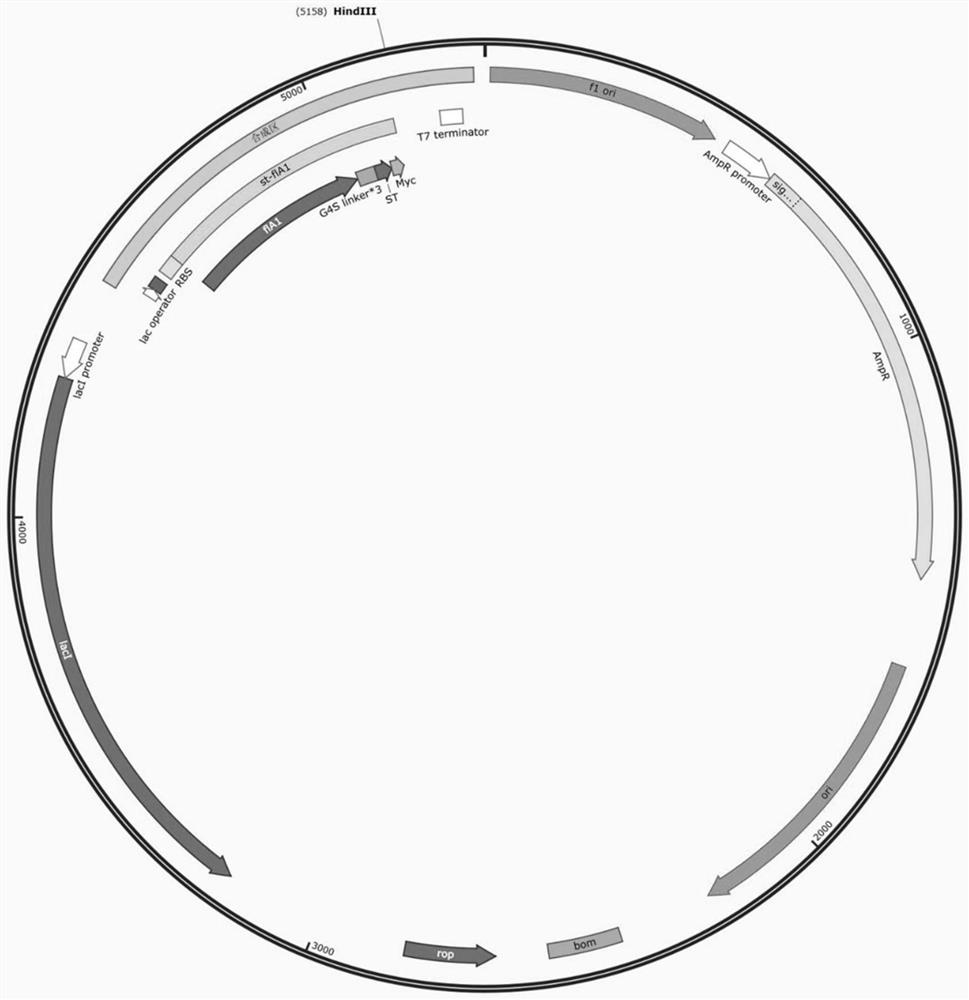
[0043] The recombinant plasmids pSQ036-TF-LP-SC and pSQ051 constructed in Example 1 (plasmid map such as image 3 shown) co-transformed into E.coliBL21 (DE3) to obtain the self-assembled SCST system strain; the pSQ036-LP-flA plasmid (see the plasmid map Figure 4 ) into E.coliBL21 (DE3) to obtain a traditional gene fusion surface display strain.
[0044] After plate activation, positive transformants were inoculated into LB medium containing 100 μg / mL ampicillin and 50 μg / mL kanamycin sulfate, and cultured on a shaker at 37°C for 12-16 hours. Draw 200uL and inoculate into liquid LB medium. Cultivate to OD at 220RPM, 37°C 600 When the absorbance value reaches 0.6-0.8, add IPTG with a final concentration of 0.5 mM and rhamnose with a final concentration of 10 mM, and culture overnight (about 22 hours) at 20° C. on a shaker.
Embodiment 3
[0045] Example 3: Identification of the surface display effect of fluorinase
[0046] (1) Preliminary verification of protein display on the cell surface by immunofluorescence staining
[0047] The specific implementation process is as follows:
[0048] 1. Measure the OD value of the bacterial strain cultivated in Example 2, and calculate the final OD 600 is 1, the dilution factor and the volume of the bacterial solution required for the final volume to be 1 mL;
[0049] 2. Centrifuge 1mL of the bacterial solution, resuspend it with PBS, take the calculated volume of the bacterial solution from the upper part, add the corresponding volume of 4% PFA (paraformaldehyde, freshly prepared) to form a 1mL system, and react for about 15 minutes;
[0050] 3. Centrifuge the reacted bacterial solution to remove PFA, add 1mL 1%-3% BSA (Bovine Serum Albumin bovine serum albumin, ready-to-use) to resuspend, and seal for 30min;
[0051] 4. Take 20uL of the reaction solution to incubate wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com