DNA coding compound library drug molecule fishing method
A drug molecule and compound library technology, applied in the field of DNA-encoded compound library drug molecule fishing, can solve problems such as the inability to meet the protein interaction mechanism and the inability to function at the level of protein interaction, and achieve sample cost savings, low cost, and high sensitivity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
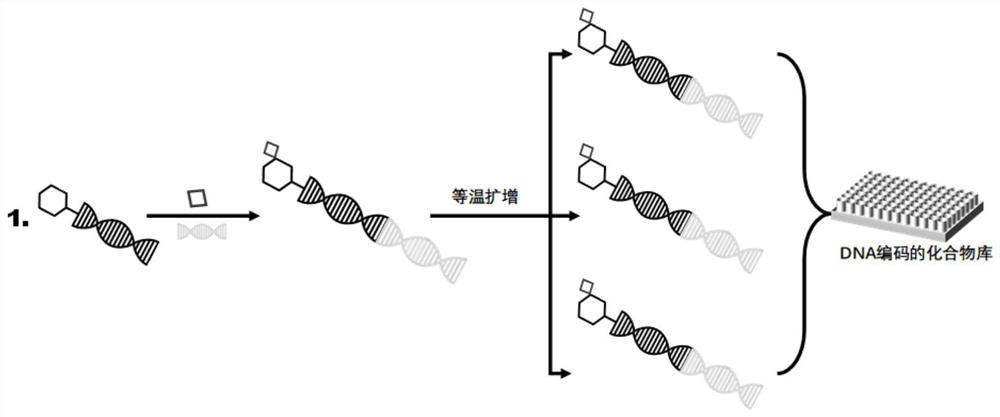
[0034] Example 1 Preparation of DNA-encoded compound library
[0035] 1. Oligonucleotide-compound ligation reaction
[0036] Dissolve a compound (1.25 μmol) in DMSO and dilute to 230 μL, add 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) (12 μL 100 mM in DMSO) and N-hydroxysuccinimide (NHS) (10 μL of 330 mM in DMSO / H2 O 2 : 1) After activation at 30°C for 20 minutes, add triethylamine (TEA) / HCl (50 μL 500 mM pH 10.0) and corresponding oligonucleotides (30 μL 500 μM) and stir overnight at 30°C. Tris-HCl (20 μL, 500 mM, pH 8.0) was added and stirred at 30° C. for 1 h to terminate the reaction, and then triethylamine acetic acid (TEAA) (500 μL, 100 mM, pH 7.0) was added. After HPLC purification, it was dried under reduced pressure, redissolved in 100 μL of H20, and then checked for concentration and quality with UV spectrophotometer and LC-ESI-MS. A total of 40 different oligonucleotide-compounds were ligated using the method described above. The structure ...
Embodiment 2
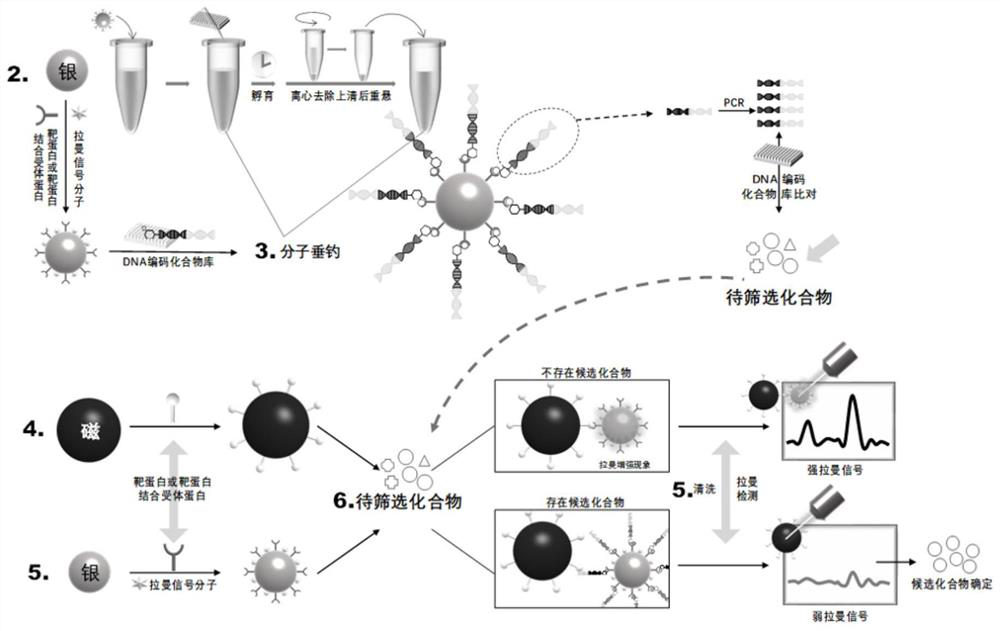
[0047] Example 2 Molecular Fishing
[0048] Synthesis of silver nano-ion probes (AgNPs): Take a 250mL three-necked flask and soak it in an acid tank overnight, take it out the next day, rinse it, and then wash it with water for injection three times to ensure that the wall is pure and free of impurities. Put in the oven to dry and cool to room temperature. During this period, 0.018 g of silver nitrate powder was accurately weighed and poured into a three-necked flask carefully. Add 100 mL of ultrapure water, shake well, connect the condenser tube and set the temperature of the electric heating mantle to 100 °C, stir and heat the solution to a slight boil, and immediately add 2 mL of 1% sodium citrate solution. Observe the color change of the solution: light yellow, dark yellow, and then gray-green, and no change in color is observed, slightly cool down to 90 ° C, continue magnetic stirring, and maintain for 40 min to obtain a silver nanoparticle dispersion system. Measure th...
Embodiment 3
[0050] Example 3 Compound structure determination
[0051] The information of the DNA fragments is determined by PCR amplification and DNA sequencing according to the oligonucleotide fragments connected to the compounds, so as to obtain the structure information of the compounds.
[0052] Take 1 μL of the supernatant as a template, add 10 μM primers (the primer structures are 5'-GCC TCC CTC GCG CCATCA GGG AGC TTG TGA ATT CTG G-3' (SEQ ID NO. 11), 5'-GCC TTG CCA GCC CGC TCA GGTAGT CGG ATC CGA CCA C-3' (SEQ ID NO. 12)) 4*2mM dNTP, 1.25U Taq enzyme and PCR buffer. The PCR steps were as follows: DNA denaturation at 94°C for 2 min, 4 cycles of 94°C for 40s, 30 cycles of 54°C for 40s, 72°C for 40s, 94°C for 40s, 64°C for 30s, 72°C for 30s, and 72°C for 7 minutes. After the reaction, the corresponding samples were mixed and purified by ion exchange column, and extracted with 50 μL QEbuffer.
[0053] The information of the DNA fragments is determined according to the results of PCR ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com