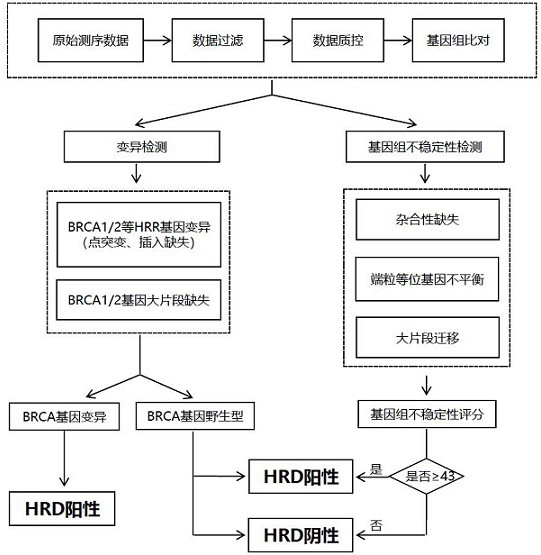
Method for constructing target set for homologous recombination repair defect detection
A technology for homologous recombination and defect detection, applied in recombinant DNA technology, biochemical equipment and methods, and microbial assay/inspection, etc., can solve the problem of increasing the potential benefit population of PARP inhibitors, large amount of sequencing data, economical cost-effectiveness It can improve the economic efficiency of detection, expand the detection rate, and reduce the detection cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
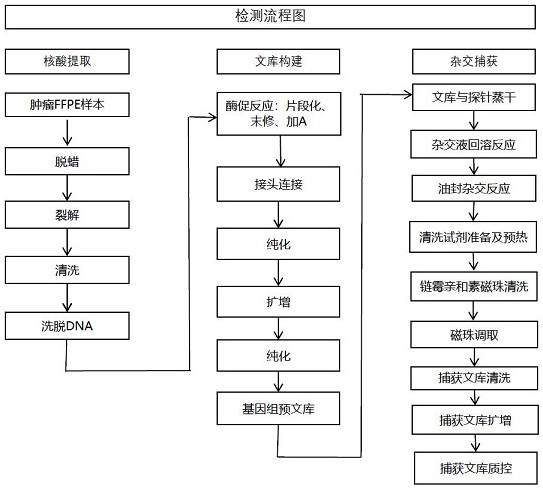
[0104] In this example, DNA is extracted from ovarian cancer paraffin-embedded (FFPE) tumor tissue samples from a clinical cooperation unit, including the following steps:
[0105] (1) Dewaxing
[0106] 1) Add 1 mL of xylene to the wax tissue, cover the tube, vortex at speed 6 for 10 seconds, incubate at 50°C for 5 minutes, centrifuge at maximum speed for 2 minutes at room temperature, and carefully suck off the supernatant with a pipette tip ;
[0107] 2) Add 1 mL of absolute ethanol to the precipitate, vortex to mix, centrifuge at the maximum speed for 2 min at room temperature, and carefully suck off the supernatant with the tip of a pipette;
[0108] 3) Store at 56°C with the lid open until the ethanol evaporates completely;
[0109] (2) cracking
[0110] 1) Add 180 μL of buffer ATL to the product of step (1) to resuspend the pellet, add 20 μL of proteinase K, vortex and mix, and incubate at 56°C for 1 h;
[0111] 2) Shake and mix, and place at 90°C for 1 hour (wait for ...
Embodiment 2
[0122] In this example, the DNA extracted from the nucleic acid in Example 1 was used. After fragmentation, adapter ligation, two rounds of purification, PCR amplification and quality control testing were performed to construct a pre-library. The steps are as follows:
[0123] (1) Enzymatic reaction
[0124] Pre-mix 5 μL Fx buffer and 10 μL Fx Enzyme Mix, store on ice, mix with 35 μL sample DNA to form a 50 μL total system, pre-cool the PCR instrument to 4°C, and set the program: 32°C, 32 min, 65°C, 30 min, 4°C Hold, hot lid 70°C, transfer the product to ice after the PCR reaction;
[0125] (2) Joint connection
[0126] Pre-mix 15 μL of water, 20 μL of buffer and 10 μL of ligase, add 5 μL of adapter stock solution, and prepare a 100 μL total system with the product of the previous step, incubate in a PCR instrument at 20°C for 15 min, and close the heat cover;
[0127] (3) Purification after connection
[0128] 1) Take the AMPure XP beads out of the refrigerator at 4°C in a...
Embodiment 3
[0138] In this example, the pre-library constructed in Example 2 is used to capture the target fragment, and the steps are as follows:
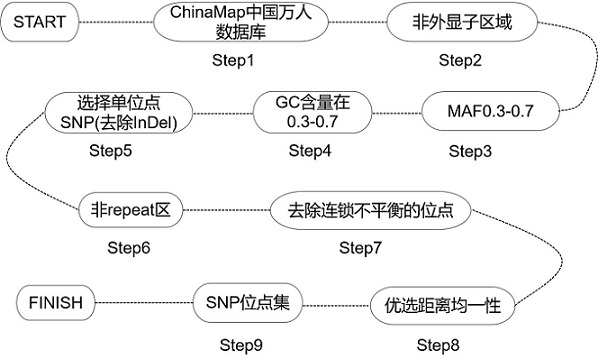
[0139] (1) Add 8 μL blockers, 5 μL blocker solution and 5 μL probe composition of the present invention to the pooled pre-library (the molar concentration ratio of HRR gene sub-probe group A to SNP site sub-probe group B is 4:1) , placed in a vacuum concentrator and evaporated to dryness at 45°C. During this period, the streptavidin magnetic beads and the purified magnetic beads can be taken out and equilibrated at room temperature for 30 min. The specific design method of the sub-probe group A is as follows:
[0140] 1) According to the UCSC database, the genome corresponding sequences of the coding regions of 40 genes were obtained, and the repeated regions were marked according to the database. The repeated regions include SINE, LINE and LTR, etc., for these repeated regions, no probe design is performed;
[0141] 2) For the gene coding s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com