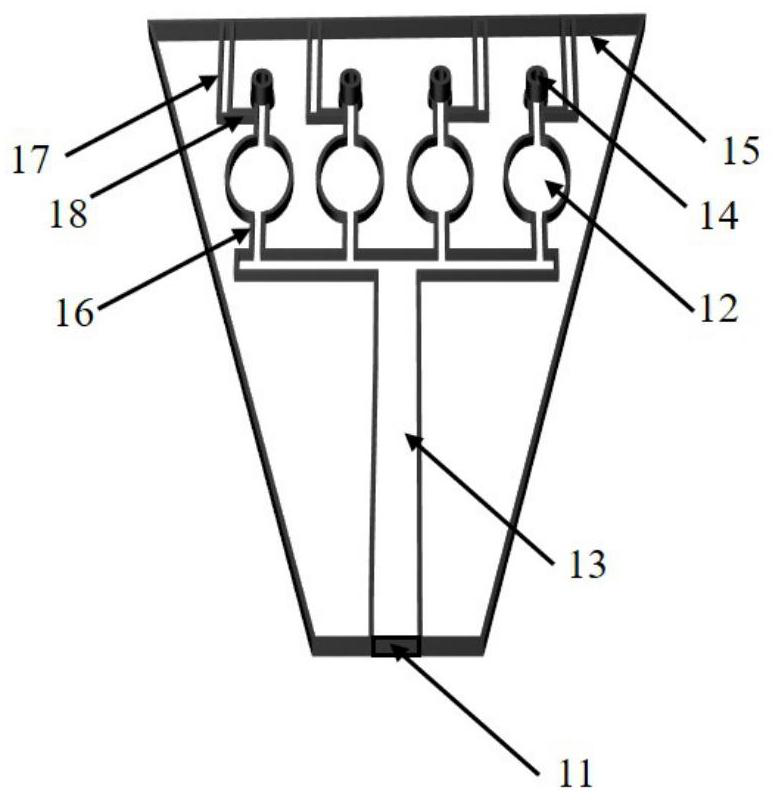
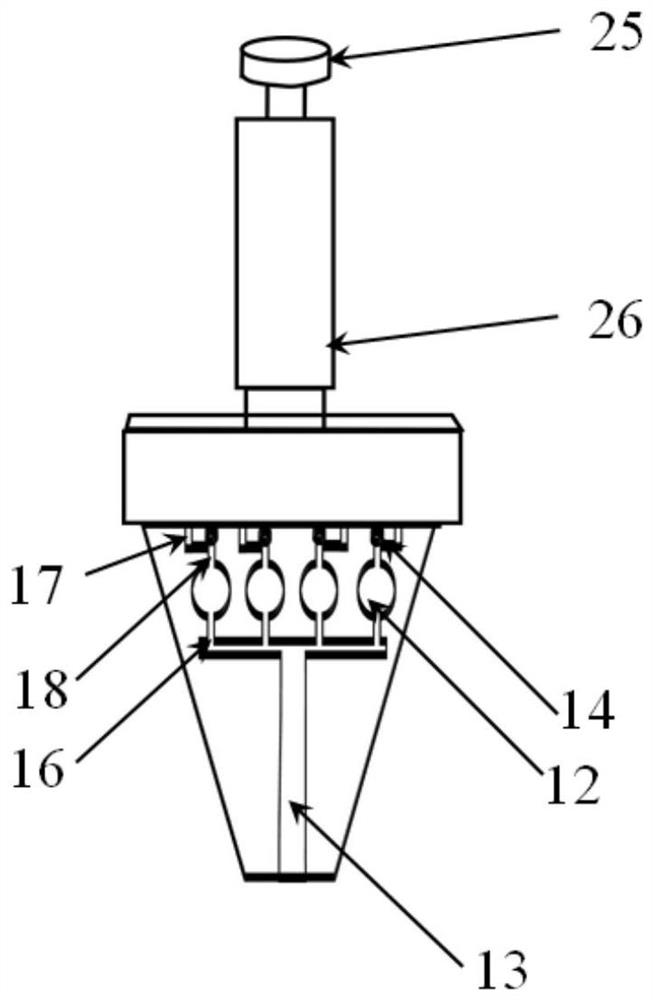
Microfluidic chip, detector, detection method and application for detecting bacteria
A technology of microfluidic chips and detectors, which is applied to chemical instruments and methods, containers used in laboratories, and analysis through chemical reactions of materials. It is unfavorable for on-site diagnosis and other problems, and achieves the effect of improving detection sensitivity, facilitating mass production, and small size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] The above method and device are used to detect the content of pathogenic Escherichia coli.
[0073] Pathogenic Escherichia coli: BNCC 133264, purchased from Beina Biological Company.
[0074] The sequence of the priming chain is shown in SEQ ID No. 1, which was synthesized by Shanghai Sangon Biological Company.
[0075] The aptamer sequence is shown in SEQ ID No. 2, which was synthesized by Shanghai Sangon Biological Company.
[0076] Reagents are H1 and H2 chains for the hybridization chain reaction, and hemin. The sequence of the H1 chain is shown in SEQ ID No. 3, and the sequence of H2 is shown in SEQ ID No. 4, both of which were synthesized by Shanghai Sangong Company. Hemin has a CAS# of 16009-13-5 and was purchased from SigmaAldrich.
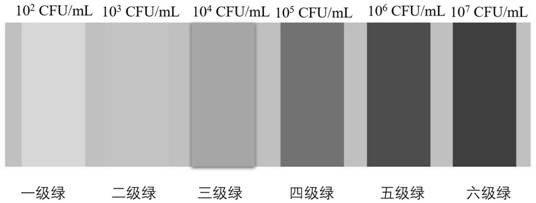
[0077] According to the detection steps described in this application, after reaching S3, it was observed that the color of the reagent in the reaction chamber reached the fifth-grade green, and the content of pathogenic E. 6 Th...
Embodiment 2
[0082] The content detection of Bacillus cereus is carried out by using the above method and device.
[0083] Bacillus cereus: BNCC 336744, purchased from Beina Bio.
[0084] The sequence of the priming chain is shown in SEQ ID No. 1, which was synthesized by Shanghai Sangon Biological Company.
[0085] The aptamer sequence is shown in SEQ ID No. 5, which was synthesized by Shanghai Sangon Biotechnology Co., Ltd.
[0086] Reagents are H1 and H2 chains for the hybridization chain reaction, and hemin. The sequence of the H1 chain is shown in SEQ ID No. 6, and the sequence of the H2 chain is shown in SEQ ID No. 7, both of which were synthesized by Shanghai Sangong Company. Hemin was the same as in Example 1.
[0087] According to the detection steps described in this application, after S3, it is observed that the color of the reagent in the reaction chamber reaches the second-level green, and the content of Bacillus cereus determined after comparing with the colorimetric card ...
Embodiment 3
[0092] The above method and device are used to detect the content of Staphylococcus aureus.
[0093] Staphylococcus aureus: BNCC 186335, purchased from Beina Bio.
[0094] The sequence of the priming chain is shown in SEQ ID No. 1, which was synthesized by Shanghai Sangon Biological Company.
[0095] The aptamer sequence is shown in SEQ ID No. 8, which was synthesized by Shanghai Sangon Biological Company.
[0096] Reagents are H1 and H2 chains for the hybridization chain reaction, and hemin. The sequence of the H1 chain is shown in SEQ ID No. 9, and the sequence of the H2 chain is shown in SEQ ID No. 10, both of which were synthesized by Shanghai Sangong Company. Hemin was the same as in Example 1.
[0097] According to the detection steps described in this application, after reaching S3, it is observed that the color of the reagent in the reaction chamber reaches the sixth-grade green, and the content of Staphylococcus aureus determined after comparing with the colorimetr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| width | aaaaa | aaaaa |
| width | aaaaa | aaaaa |
| width | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com