SARS-CoV-2 S protein specific antibody or fragment thereof and application thereof
A sars-cov-2 and sars-cov-2s technology, applied in the field of antibody engineering, can solve the problems of monoclonal antibodies with no sequence structure, vaccine and therapeutic antibody research difficulties, and achieve high clinical application potential and high affinity , The effect of clear amino acid sequence structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1: Recombinant expression of SARS-CoV-2 antigen and host receptor
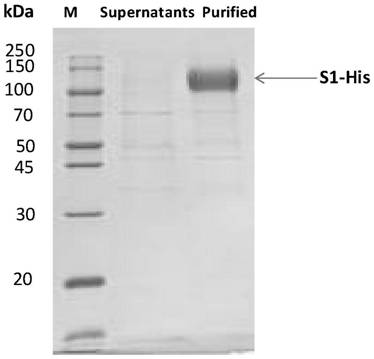
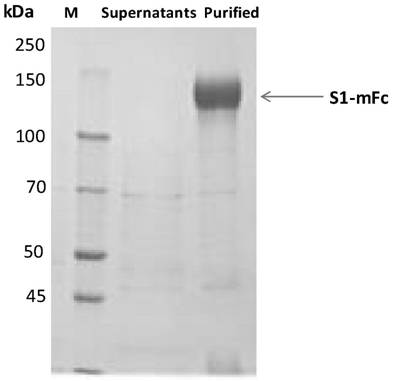
[0061] The fully synthesized gene S1RBD (Accession: QHD43416.1, 319-541aa) was cloned into eukaryotic transient expression vectors with His tag or mFc tag at the C-terminus by enzyme digestion, and the obtained expression plasmid was transformed into Escherichia coli was amplified, the S1RBD-his and S1RBD-mFc expression plasmids were isolated, and according to the instructions of the transfection reagent 293fectin (Cat: 12347019, Gibco), the plasmids were transferred into HEK293 cells for recombinant expression. 5-6 days after cell transfection, the culture supernatant was taken, and S1RBD-mFc was purified by ProA affinity chromatography column to obtain S1RBD-mFc protein. S1RBD-his was purified by HisTrap HP column affinity chromatography to obtain S1RBD-his protein. And the obtained recombinant protein was tested for purity by SDS-PAGE ( Figure 1-2 ). The coding nucleic acid sequence of S1...
Embodiment 2
[0064] Example 2: B cell screening to obtain anti-new coronavirus S1RBD specific antibody
[0065] Using FITC-S1RBD-hFc as an antigen, the specific memory B cells of recovered patients with novel coronavirus infection were sorted. Single-cell PCR technology (method and primer reference New Biotechnology, 2010. Volume 27, Number 2, P110-117 and pages 114 to 117 in the book Human Monoclonal Antibodies) amplified antibody light and heavy chain genes, and performed sequencing analysis. The correctly sequenced antibody light and heavy chain variable region genes were synthesized and cloned into the transient expression vector pKN019 containing the light chain constant region and the transient expression vector pKN041 containing the heavy chain constant region IgG1 gene, and the HEK293 system was used for transient expression and purification to obtain recombinant antibody MW07 .
[0066] The method for single-cell PCR amplification of antibody light and heavy chain genes is as fol...
Embodiment 3
[0102] Example 3. MW07 and S1RBD affinity
[0103] The Fortebio protein interaction system was used to determine the binding affinity of the antibody to the S1-RBD protein antigen. The main steps are as follows:
[0104] Using Fortebio's Octet QKe system, the anti-human antibody Fc fragment capture antibody (AHC) bioprobe was used to capture the binding and dissociation changes of different concentrations of the assay antibody to the monovalent S1RBD. During the measurement, four different concentrations of MW07 (45nM, 30nM, 15nM, 7.5nM) flowed over the surface of the AHC probe (Cat: 18-5060, PALL) for 120s. S1RBD-His diluted in different concentrations was used as mobile phase. The binding time is 300s and the dissociation time is 300s. After the experiment was completed, the response value of the blank control was deducted, and the 1:1 Langmuir binding model was fitted with software to calculate the kinetic constant of antigen-antibody binding. The result is as Figure ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| absorbance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com