Dna-cutting agent
A technology of DNA molecules and DNA sequences, applied in the field of new Cas nucleases, which can solve problems such as restricting the use of nucleases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1. Testing the activity of DfCas9 protein in cleaving various DNA targets
[0064] To test the ability of DfCas9 to recognize different DNA sequences flanked by the motif 5'-NN(G / A)NA(C / T)N-3', experiments with in vitro cleavage of DNA targets from the human grin2b gene sequence were performed ( See Table 1).
[0065] Table 1. DNA targets isolated from the human grin2b gene.
[0066] DNA target PAM attttctctcattctgcaga gcaaata tctaagacaggttacgtgat gtagatc aatgaaaggagataaggtcc ttgaatt caccttttattgccttgttc aaggatt ggcattgctgtcatccctcgt gggcact ttgcagtatctagcctcttc taagaca
[0067] Perform in vitro DNA cleavage reactions under similar conditions to previous experiments. Guide crRNAs were synthesized for each target sequence. A fragment of the human grin2b gene with a size of approximately 760 bp was used as the DNA target:
[0068] .
[0069] A sequence flanked from the 3' end by a DNA region correspo...
Embodiment 2
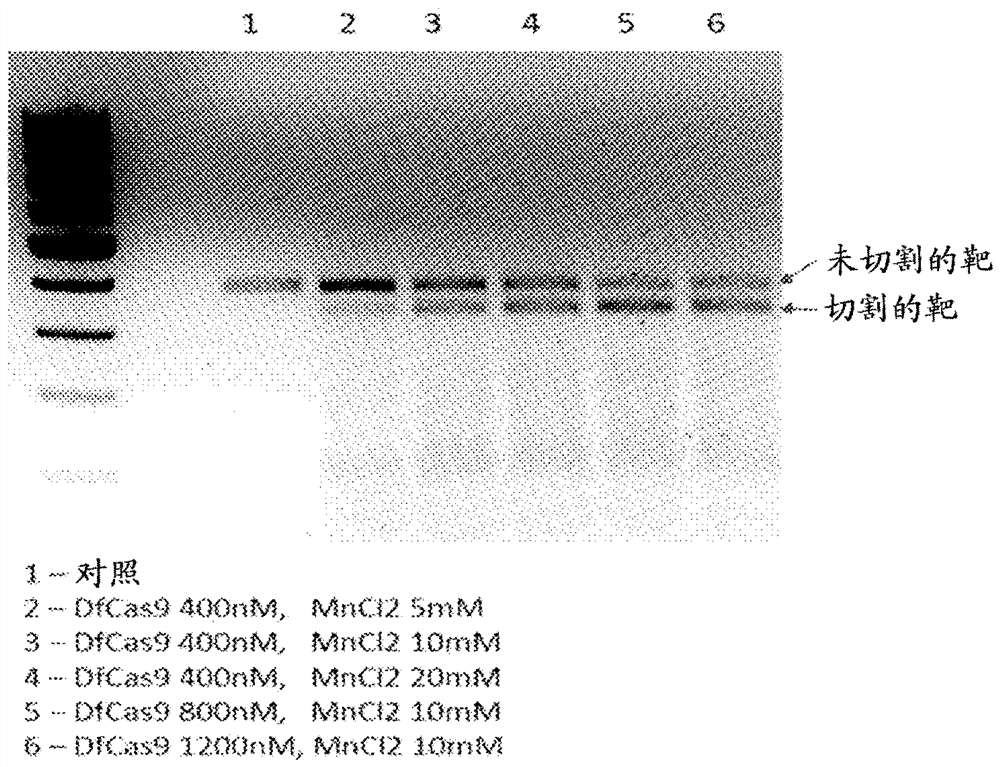
[0071] Example 2. Effect of Mn2+ ion concentration on nuclease functionality
[0072] To test the effect of Mn2+ ions on DfCas9 nuclease activity, experiments were performed in vitro to cleave double-stranded DNA fragments containing target DNA sequences restricted to the PAM sequence AAAAAC with the consensus sequence 3'-NN(G / A) NA(C / T)-5'. Reactions were performed using 1x CutSmart Buffer (NEB), target DNA at a concentration of 20 nM, and trRNA / crRNA at a concentration of 2 μM.
[0073] As a DNA target, we use
[0074] .
[0075] React using tracrRNA:
[0076] and
[0077] .
[0078] Figure 7 It was shown that increasing the concentration of MnCl2 from 5 mM to 10 mM resulted in more efficient cleavage of the DNA target, whereas further increases in the concentration of divalent ions did not affect the reaction efficiency. Therefore, efficient DfCas9 activity requires the presence of manganese ions at a concentration of 10 mM or higher.
Embodiment 3
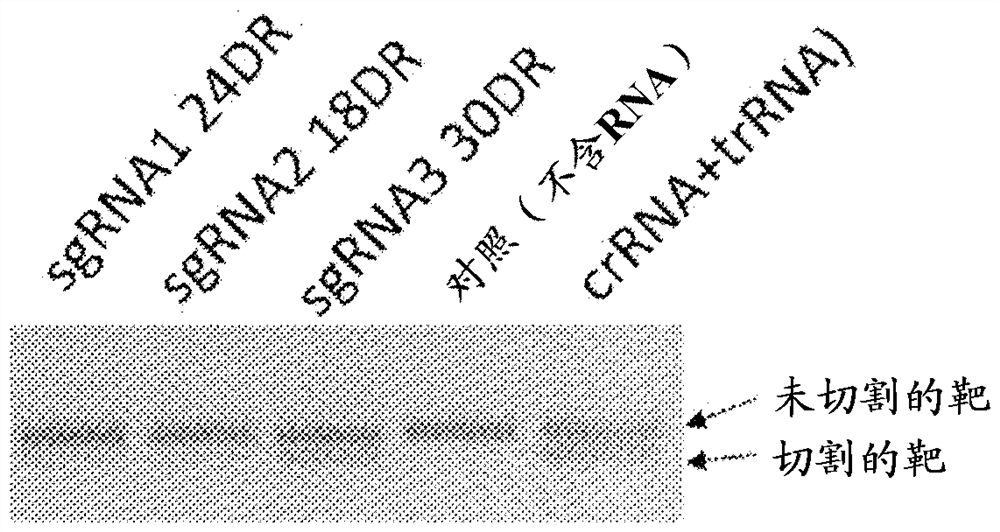
[0079] Example 3. Use of hybrid guide RNAs for cleavage of DNA targets
[0080] sgRNA is a form of guide RNA that is a fusion of tracrRNA (tracer RNA) and crRNA. To select the optimal sgRNA, we constructed three variants of this sequence that differ in the length of the tracrRNA-crRNA duplex. RNA was synthesized in vitro, and experiments were performed on it to cleave DNA targets ( Figure 8 Reactions for DNA cleavage by DfCas9 using various sgRNA variants are shown).
[0081] The selected sgRNA3 was as efficient as native tracrRNA and crRNA sequences, cleaving more than 50% of the DNA target.
[0082] While modifying the sequence that pairs directly with the DNA target, this sgRNA variant can be used to cleave any other target DNA.
[0083] The following RNA sequences were used as hybrid RNAs:
[0084] 1-sgRNA1 24DR:
[0085]
[0086] 2 - sgRNA2 18DR
[0087]
[0088] 3 - sgRNA3 30DR
[0089]
[0090] 4 - Control (without RNA)
[0091] 5 - Positive control (c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com