DNA (deoxyribonucleic acid) bar code and primer group for identifying source area of champignon luteo-virens and application of DNA bar code and primer group
A technology of origin and primer set, which is applied in the direction of recombinant DNA technology, DNA/RNA fragments, microbial determination/inspection, etc., can solve the problems of difficulty in popularization, poor reliability and repeatability of results, and high cost, and achieve the goal of overcoming inaccuracy Time-consuming and labor-intensive, stable and reliable results, and short detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] The construction of the DNA barcode of embodiment 1 yellow-green mushroom
[0067] Genome sequencing was carried out on the samples of Dangxiong County in Tibet Autonomous Region, Qilian County in Qinghai Province, and Shiqu County in Sichuan Province. The SSR loci in the genome sequences were analyzed using the MISA program.
[0068] Design primers for PCR amplification of these SSR sites, keep primers that can amplify the corresponding fragments, and discard invalid primers
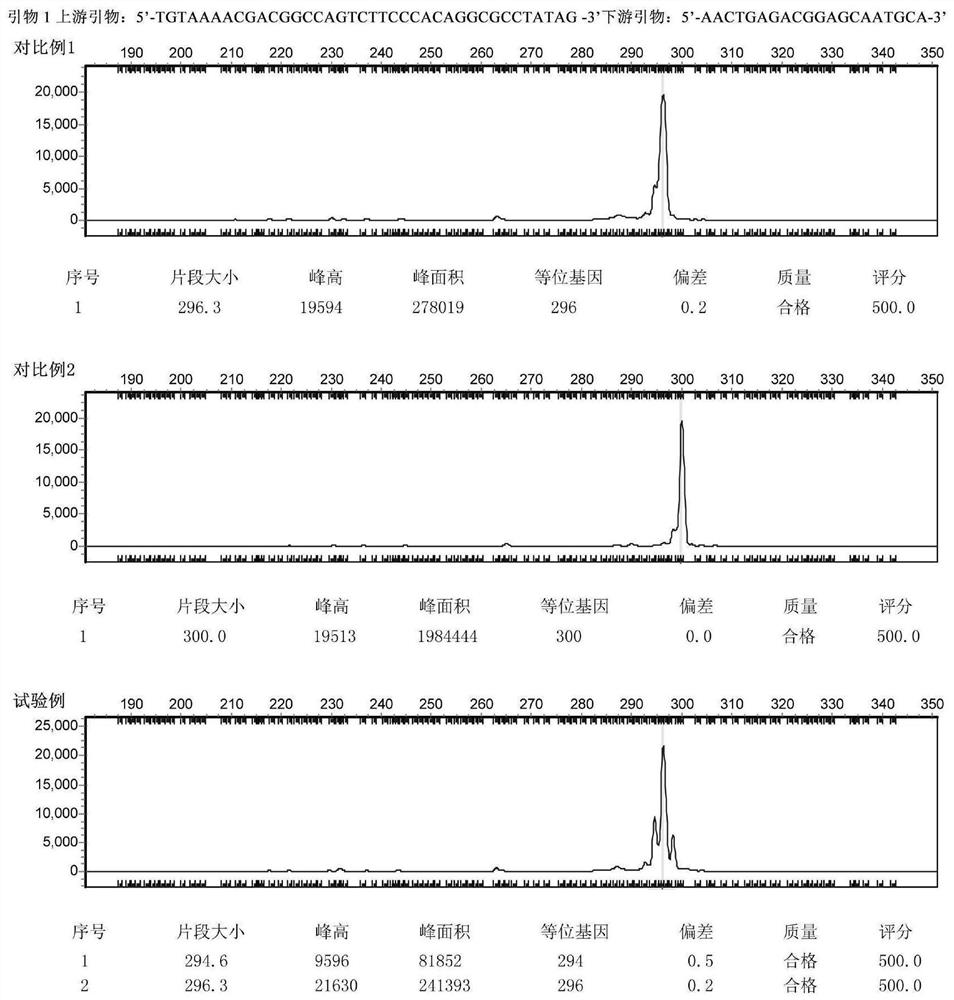
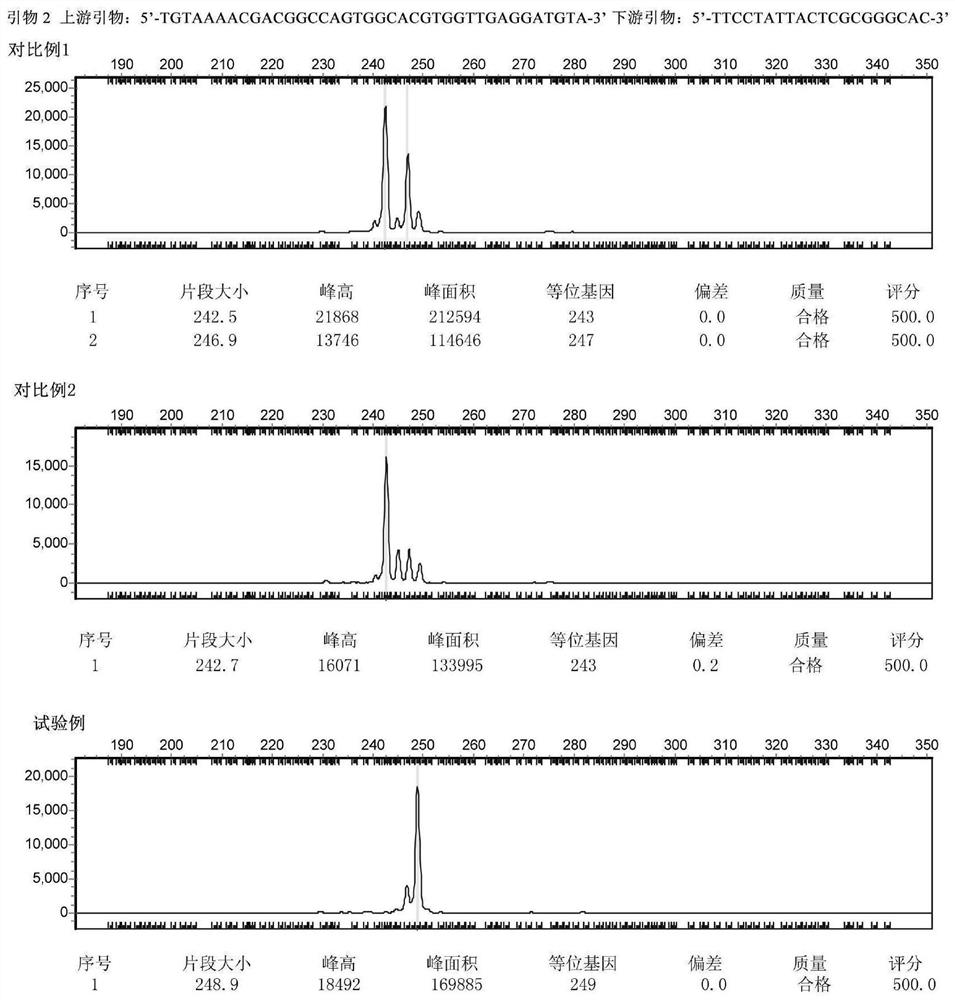
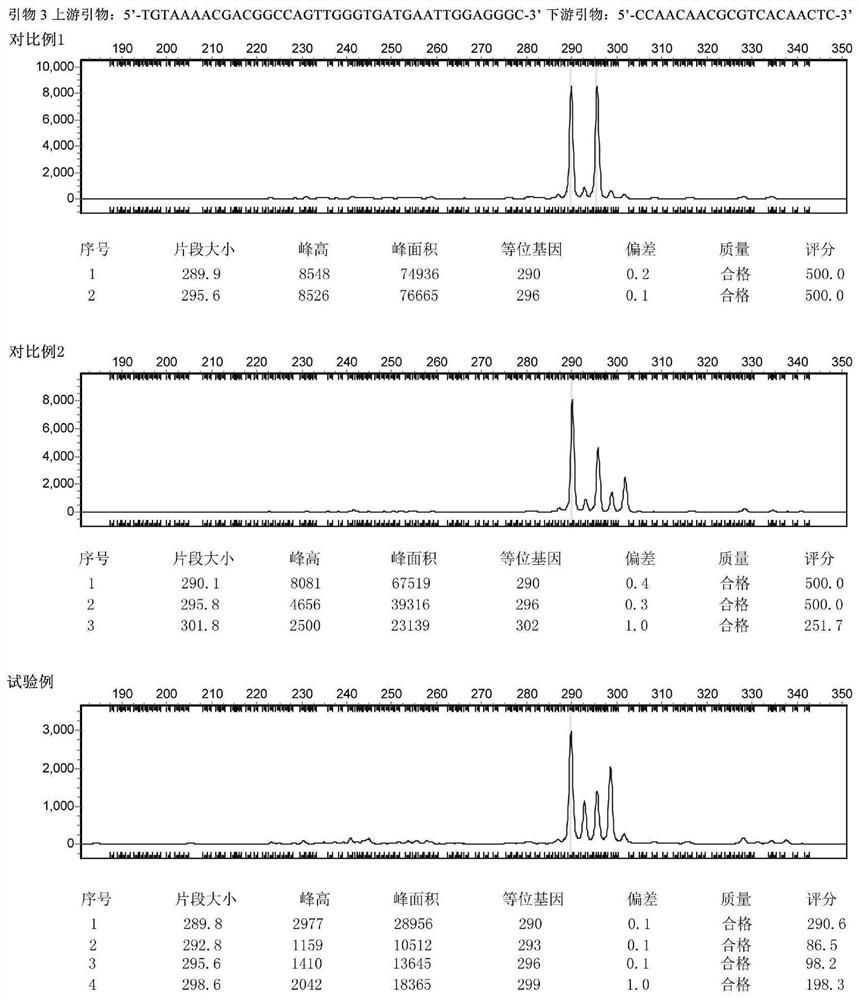
[0069] The samples from the above three origins were respectively amplified using effective primers and detected by capillary electrophoresis. After analysis, the corresponding simple sequence repeat (SSR) locus was established to distinguish the origin. Finally, 4 pairs of primers were obtained (see Table 1), and the fragment polymorphism obtained by using these 4 pairs of primers to amplify the sample genome can assist in the identification of the origin of the yellow-green mushroom.
[0070]...
Embodiment 2
[0072] Embodiment 2 Identification of the place of origin of yellow-green mushroom
[0073] (1) Use the Sangon Bioengineering (Shanghai) Co., Ltd. Ezup Column Fungal Genomic DNA Extraction Kit (Cat. No. B518259) to extract the genome of the yellow-green mushroom sample, and dilute it to 20 ng / μL for fluorescent PCR amplification.
[0074] (2) Using the primers in Table 1 to amplify the SSR DNA barcode by fluorescent PCR, the origin of the yellow-green Pleurotus edodes can be identified by reading the information of the DNA barcode.
[0075] Fluorescent PCR amplification reaction system (10 μL): 2×Taq PCR Master Mix 5 μL, template (genome DNA) 1 μL, upstream primer 0.1 μL, downstream primer 0.4 μL (concentration of both upstream and downstream primers is 10 uM), fluorescent M13 primer (concentration 10uM) 0.4 μL, dilute to 10 μL with sterile deionized water;
[0076] Reaction conditions: pre-denaturation at 95°C for 3 minutes; denaturation at 95°C for 30 s, drop PCR annealing ...
Embodiment 3
[0122] Example 3 Yellow-green Pleurotus pubescens origin identification DNA barcode verification
[0123] Validation of DNA barcodes for identification of the origin of Pleurotus edulis by blind test.
[0124] In the first blind test, samples were taken from Qilian County of Qinghai Province (number of origin 1-16), Shiqu County of Sichuan Province (number of origin 17-32) and Damxung County of Tibet Autonomous Region (number of origin 33-48) 16 samples were blind tested;
[0125] The second step test utilizes primers (SEQ ID NO: 1, SEQ ID NO: 2, SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 11, SEQ ID NO: 12, SEQ ID NO: 18, SEQ ID NO: 19) was amplified and subjected to capillary electrophoresis. Primer sets can be amplified using one or more pairs of combinations to distinguish blind samples and identify samples of origin with DNA barcode features;
[0126] The third step is unblinding. The results are shown in Table 3. All 16 samples from Qilian County in Qinghai Province, Shiqu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com