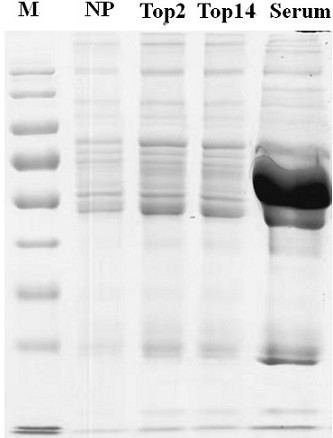
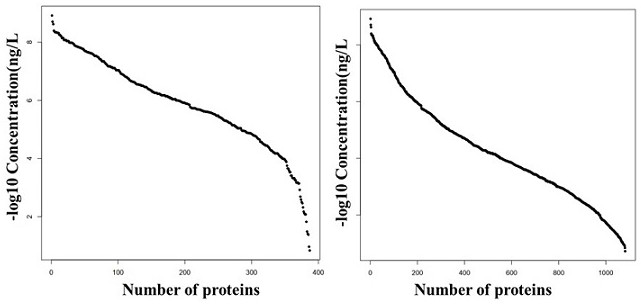
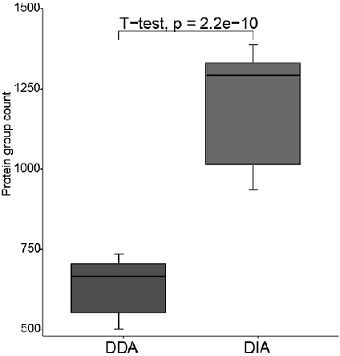
Serum protein preparation method and serum proteome mass spectrometric detection method
A serum protein and proteome technology, which is applied in the preparation of test samples, measurement devices, and material analysis by electromagnetic means, can solve the problems of increasing the scanning time of mass spectrometry detection, reducing the dynamic range of serum samples, and reducing quantitative accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0075] The invention provides a sample preparation method of serum proteome, comprising the following steps:
[0076] (1) After the serum sample is centrifuged at low temperature and low speed, the bottom particles and insoluble matter are removed, and the supernatant is retained;
[0077]The serum is 25µl to 1000µl, the low temperature is 2~8°C, preferably 4°C, the centrifugal force is 500~2000 RCF, preferably 1000RCF, and the centrifugation time is 10~20 minutes, preferably 15 minutes;
[0078] (2) Dilute the supernatant with equal volume of PBS buffer, add the mixed magnetic nanoparticles with hydrophilic and hydrophobic surface modification, and incubate at 37°C for 5-10 minutes, the mixed magnetic nanoparticles with hydrophilic and hydrophobic surface modification are Surface dextran-modified hydrophilic magnetic nanoparticles and surface polystyrene-modified hydrophobic magnetic nanoparticles total 1.25 mg, and the mass ratio is 10:1;
[0079] (3) After the serum mixed ...
Embodiment 1
[0084] Embodiment 1, the preparation of serum proteome
[0085] (1) Thaw 25 μl of serum from the refrigerator at room temperature, centrifuge at 1500 RCF for 10 minutes at 4°C, remove the bottom cells and pellets, and transfer the supernatant to a new tube.
[0086] (2) Add 25 μl PBS buffer to the supernatant, then add 1.25 mg mixed nanoparticles (mixed hydrophilic and hydrophobic magnetic nanomaterials at a mass ratio of 10:1) and incubate at 37°C for 10 minutes with rotation.
[0087] (3) After the incubation was completed, magnetic separation was performed for 5 minutes to remove the supernatant solution, the precipitate was washed with 200 μl PBS vortex shaking for 2 minutes, and the supernatant solution was removed by magnetic separation for 5 minutes.
[0088] (4) Add 20 μl of 1% DDM, 10 mM TECP, 40 mM CAA and 100 mM TEAB digestion buffer (pH8.5), react at 95°C for 10 minutes, add sequencing grade trypsin 1ug, and react at 37°C for 16 hours.
[0089] (5) Add 20% TFA (vo...
Embodiment 2
[0090] Example 2, mass spectrometry detection of serum proteome
[0091] The preparation method of the serum proteome sample is the same as that in Example 1.
[0092] 10 μl of loading buffer (0.1% (v / v) formic acid) was used to dissolve, and then 2 μl of mass spectrometry was taken for detection, and the data was collected using ThermoScientific nanoliter liquid chromatography tandem high-resolution mass spectrometry (nLC-Easy1200-OrbitrapExploris 480).
[0093] The specifications of nanoliter liquid chromatography pre-column and analytical column are as follows:
[0094] Pre-column: 3µm particle size C 18 Filler, 2cm×150µm inner diameter (filler is Dr.Maisch GmbH company).
[0095] Analytical column (separation column): 1.9µm particle size C 18 Filler, inner diameter 25cm×75µm (filler is Dr.Maisch GmbH company).
[0096] Mobile phase A is 0.1% (v / v) volume ratio (1000ul of water is added to 1ul of formic acid) formic acid (formic acid, FA) aqueous solution, mobile phase ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Centrifugal force | aaaaa | aaaaa |
| Particle size | aaaaa | aaaaa |
| Column length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com