Novel protein biotin ligase and proximity labeling system PhastID based on same
A biotin ligase, protein labeling technology, applied in the field of adjacent labeling systems, can solve the problems of weak labeling ability, long time required, large molecular weight of enzyme protein, etc., and achieve short labeling time, high labeling intensity and great application prospects. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
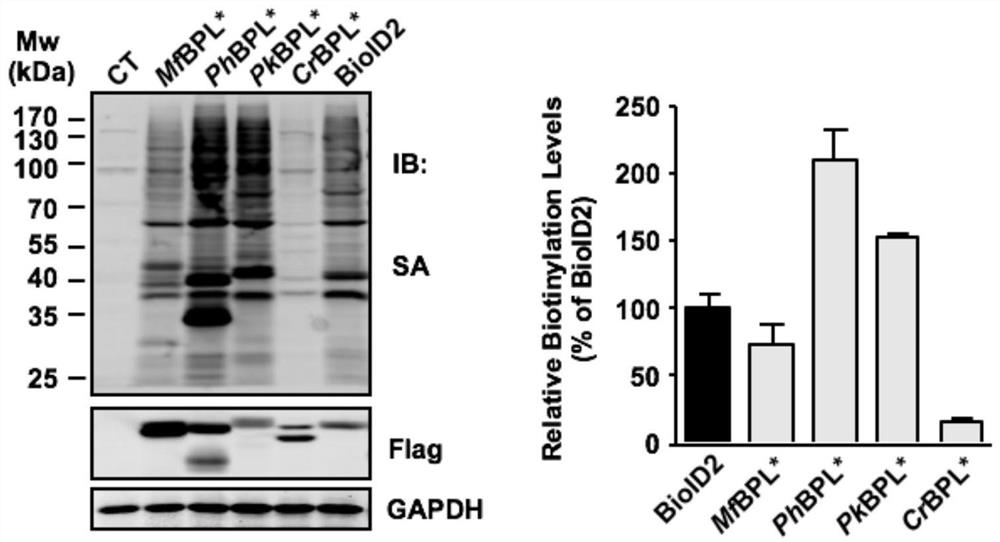
[0039] Example 1 Obtaining of protein biotin ligase BPL* (PhBPL*, PkBPL* and MfBPL*)
[0040] Obtain biotin ligase sequences of various species on NCBI, including the BPL derived from the archaea Pyrococcushorikoshii, called PhBPL, whose amino acid sequence is shown in SEQ ID NO.1; the BPL derived from the archaea Pyrococcus kukulkanii, called PkBPL, whose amino acid sequence is shown in SEQ ID NO.2; and the BPL derived from the archaea Methanocaldoccus fervens, called MfBPL, whose amino acid sequence is shown in SEQ ID NO.3; analyze its GNGR motif (N represents a basic amino acid ), and mutate the above protein into GNGG form; PhBPL R48 amino acid residue mutation: R48G or R48S mutation, namely PhBPL (R48G) or PhBPL (R48S), called PhBPL*; PkBPL R48 amino acid residue mutation: R48G or R48S mutation , that is, PkBPL (R48G) or PkBPL (R48S), called PkBPL*; MfBPL R40 amino acid residue mutation: R40G, that is, MfBPL (R40G), called MfBPL*.
[0041] The coding nucleotide sequences...
Embodiment 2
[0054] Example 2 Construction and expression of protein biotin ligase BPL* recombinant plasmid
[0055] (1) Construction of recombinant plasmids
[0056] The eukaryotic expression vector pcDNA3.1(-) was recovered after double digestion with BamHI and HindIII, and then combined with the novel biotin ligase BPL* (PhBPL*, PkBPL* and MfBPL*) recovered after double digestion with BamHI and HindIII PCR fragments (SEQID NO.4~6) were ligated under the action of T4 ligase, transformed into DH5α competent cells, spread on LB plates containing ampicillin, picked single colonies after overnight culture, and extracted plasmids for sequencing identification , the identified positive plasmid is the desired recombinant plasmid.
[0057](2) Transient expression of novel biotin ligase in mammalian cells
[0058] In order to construct a mammalian cell line expressing a novel biotin ligase, the pcDNA3.1 recombinant plasmid in step (1) was transfected into 293T cells using the Lipofectamine 2000...
Embodiment 3
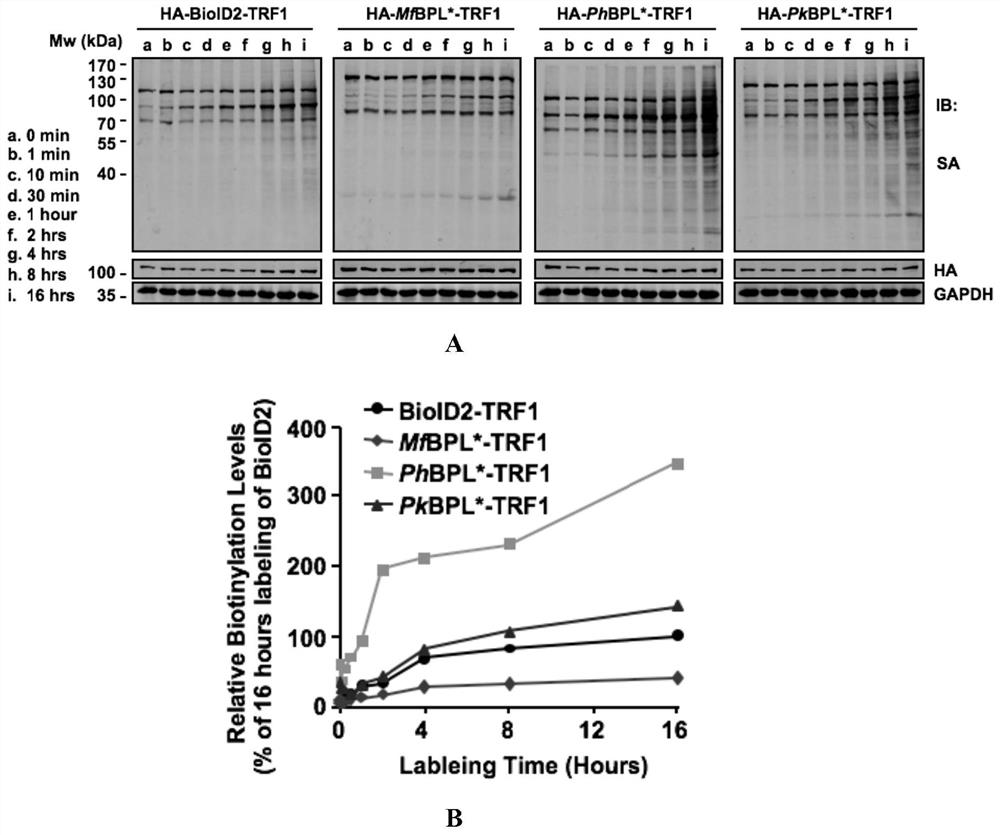
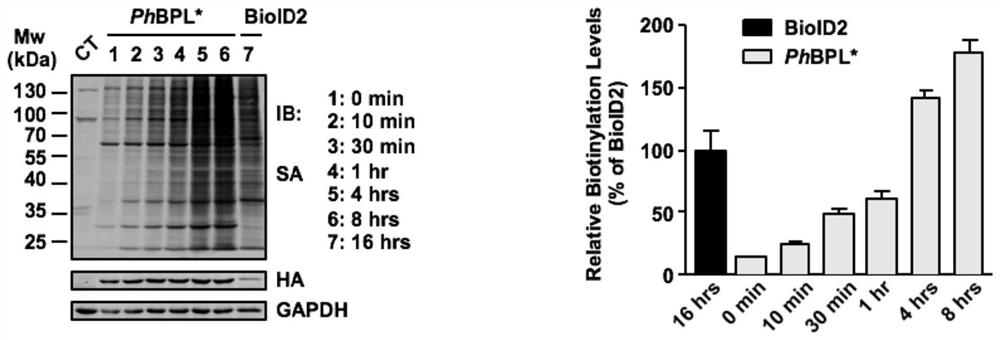
[0079] Example 3 Exploration of conditions for labeling adjacent proteins with protein biotin ligase BPL*
[0080] The novel biotin ligases BPL* (PhBPL*, PkBPL* and MfBPL*) (SEQ ID NO.4-6) and the telomere-binding protein TRF1 were recombined and cloned into the lentiviral vector respectively, and the HA antigen was added to its N-terminus , forming the pLenti-HA-BPL*-TRF1 vector, and constructing the pLenti-HA-BPL*-NLS vector at the same time.
[0081] Spread HEK293T cells to 6cm 2 Petri dish, replace its density to 70%-80% for transfection. Before transfection, select and prepare the following transfection systems shown in Table 3 as needed:
[0082] Table 3 Transfection system
[0083]
[0084] After the system is prepared, blow and mix with a pipette gun, and let it stand at room temperature for 15 minutes. Add it to the cultured cell culture medium, shake it back and forth to mix well; change the fresh medium after 6 hours, and continue to cultivate until 48 hours;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com