Single cell RNA sequencing
A single-cell, cell-based technology used in molecular biology and drug development to solve problems such as slowness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Drug treatment, scRNA-seq, and sequencing data analysis
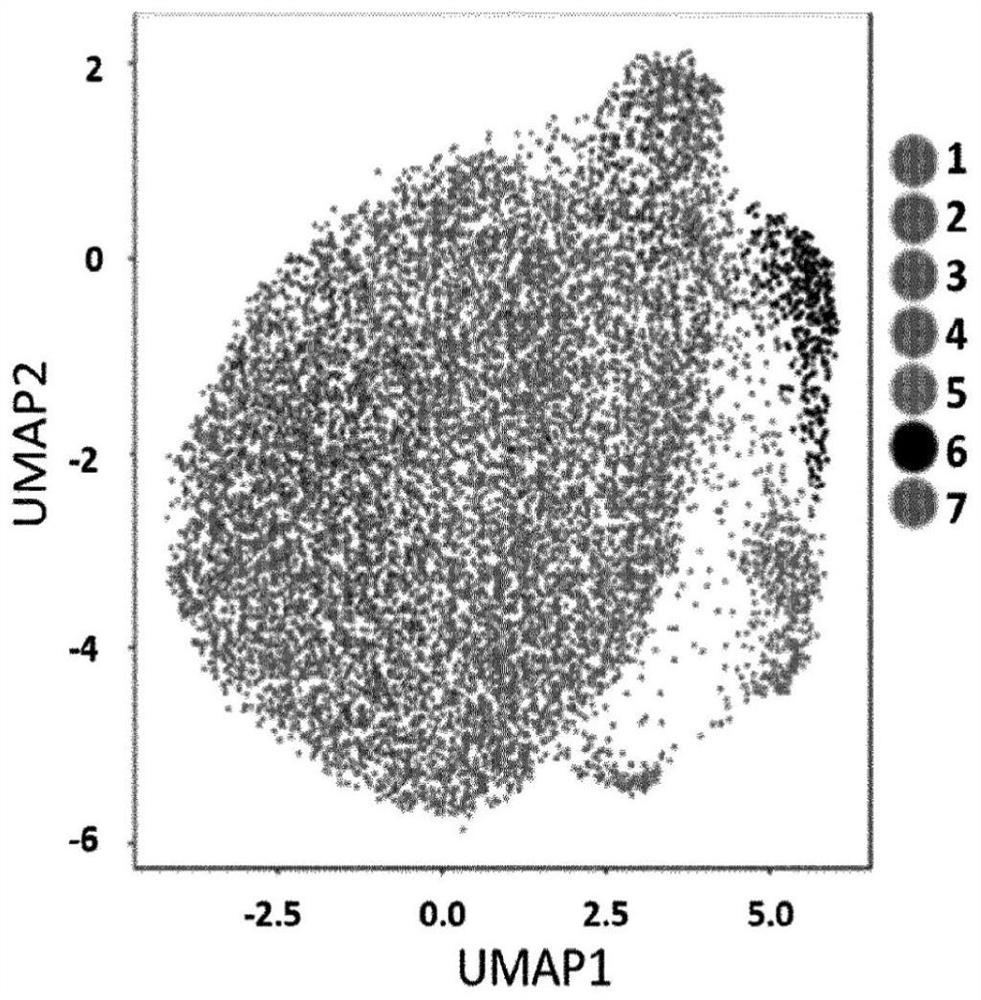
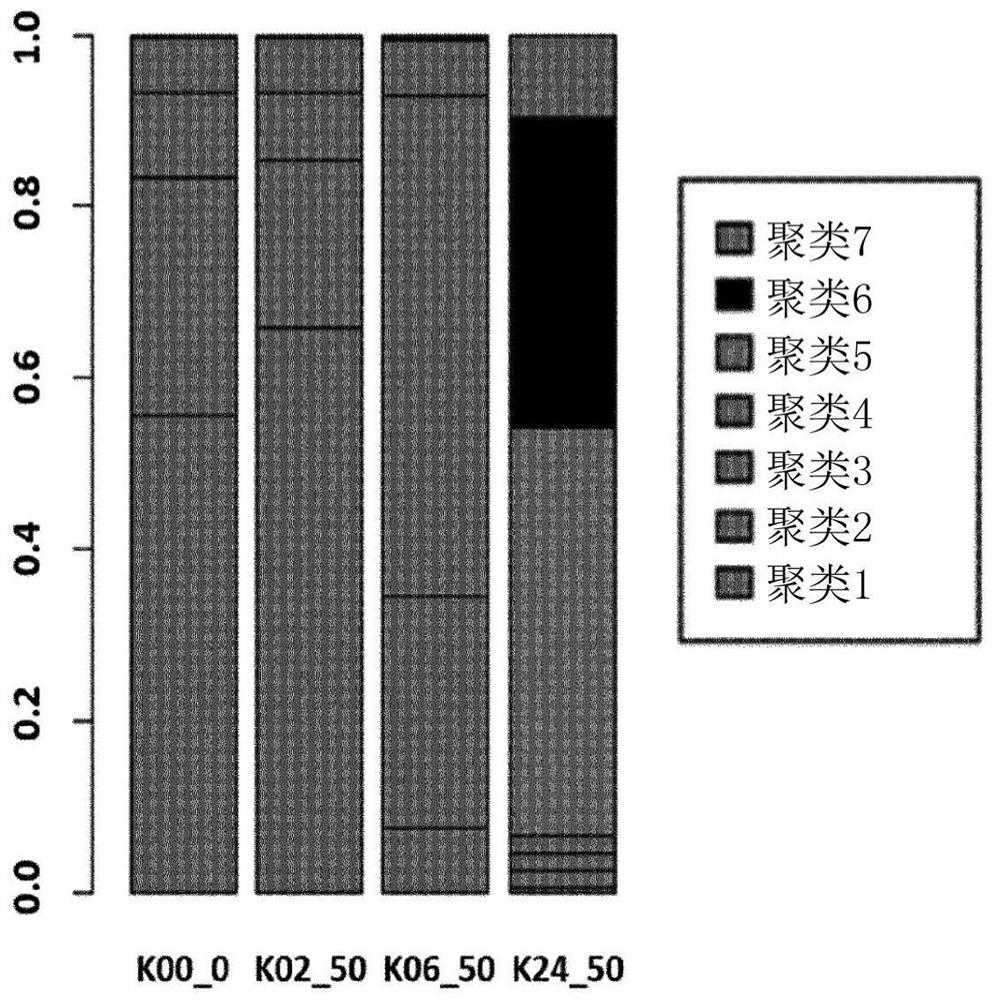
[0078] K562 cells were cultured in medium and then exposed to 50 nM oxaliplatin. After 0, 2, 6, and 24 hours of exposure, cells were washed and resuspended in PBS, and samples were denoted as K00_0, K02_50, K06_50, K24_50. Through single cell 3' gel beads, Microfluidic chips and library kits handle single cells. Library sequencing was performed on an Illumina sequencer.
[0079] Compositional changes of cell line populations are visualized by UMAP plots. Cells were subjected to Gene Set Enrichment Analysis (GSEA).
[0080] Total RNA was isolated using the AllPrep DNA / RNA Mini Kit (Qiagen, Cat. No. 80204), and measured using a Qubit2.0 Fluorometer (Life-Technologies, CA, USA) and a NanoPhotometer Spectrophotometer (IMPLEN, CA, USA).
[0081] Prepare a concentration of 1 x 10 5 single cell suspension in PBS (HyClone) per mL. Then load the single cell suspension to the microfluidic device, according to the n...
Embodiment 2
[0089] Drug handling, scRNA-seq, and sequencing data analysis
[0090] K562 cells were cultured in medium and then exposed to different concentrations (0 μM, 5 μM, 20 μM) of lysine-specific histone demethylase 1A (LSD1) inhibitors for 0, 6, 24 hours. Cells were washed and resuspended in PBS, and samples were denoted as LSD1_0_0, LSD1_6_0, LSD1_6_5, LSD1_6_20, LSD1_24_0, LSD1_24_5, and LSD1_24_20. Through single cell 3' gel beads, Microfluidic chips and library kits handle single cells. Library sequencing on an Illumina sequencer
[0091]Compositional changes in cell line populations are visualized by t-SNE plots. GSEA was performed on cells considering different datasets (Gene Ontology database and KEGG signaling pathway database) by cluster profiler. Pseudotime trajectories for each single sample were identified by using Monocle2 under default parameters. Pseudo-time trajectories for all cell lines were analyzed, along with the position of each clustered cell on the tra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com