Search for cancer markers by a novel screening method
a cancer marker and screening method technology, applied in the field of peptide concentration methods, can solve the problems of low specificity of pancreatic cancer markers, unsatisfactory treatment results of pancreatic cancer, and low usefulness in early diagnosis, and achieve the effect of efficient screening novel pancreatic cancer markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of a Serum-free Culture Supernatant
[0099] A cell line was cultured to confluence in a RPMI1640 medium containing 10% fetal bovine serum (Life Technologies). After aspirating the medium, it was washed three times in 10 ml each of a serum-free medium, and further cultured at 37° C. for 1 hour. After two more washings, 3 ml of the medium per 10 cm culture dish was added and cultured for 48 hours.
[0100] After removing the cells in a centrifuge, the culture supernatant was transferred to a polypropylene tube and stored at 4° C. until analysis. The protein concentration was determined using the Protein Assay kit (Bio-Rad) with immunoglobulin as a standard. As a control, a medium containing 10% serum was added to an unused dish and cultured for 72 hours, which was then washed as described above and used. The medium obtained is termed a “cell-free” culture supernatant. For all cell lines, trypan blue dye exclusion test was performed, confirming that 95% or more cells survived ...
example 2
Optimization of SELDI Analysis for Profiling Low Molecular Weight Proteins
[0101] Mass was determined using Ciphergen SELDI Protein Biology System 2 (Ciphergen Biosystems, Inc., CA) as a mean of 110 laser shots. For spectral analysis, monovalent and divalent molecular ions of bovine insulin were used to carry out external calibration (m / z 5734.56 and 2867.78). Values of m / z when referred to in the text were rounded to three-place or four-place significant figures. Precision of mass was better than 1000 ppm (0.1%) in the analytical range.
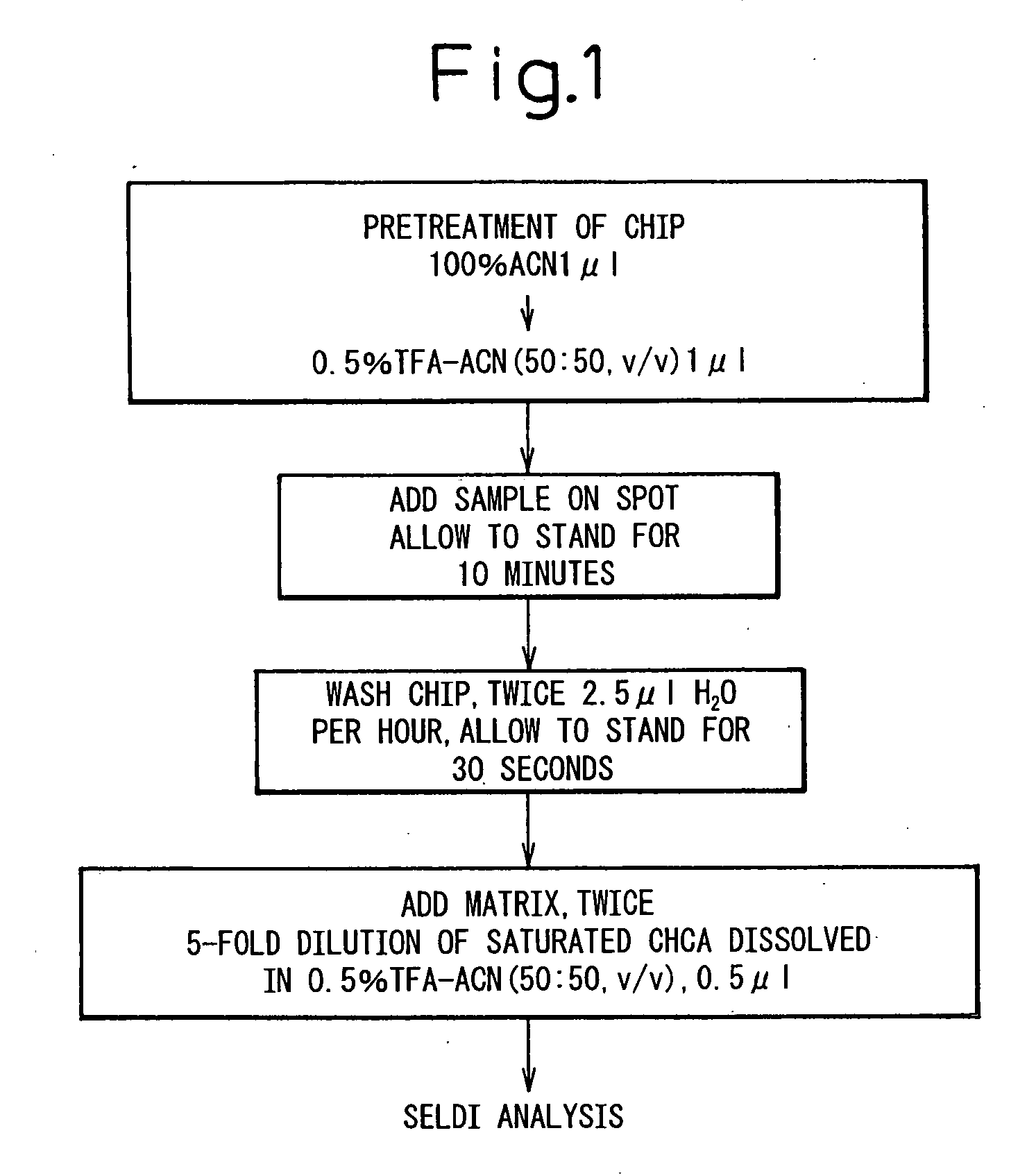
[0102] In order to analyze low molecular weight proteins, the H4 protein chip array having a chemical surface that captures protein with hydrophobic interaction was used. The experimental conditions for sample binding and matrix cocrystals were optimized to obtain spectra. As the application of mass spectrometry in protein profiling of biological materials is at a development stage and thus there are no established protocols, the optimization thereo...
example 3
Comparison of Tricine Electrophoresis and SELDI Analysis
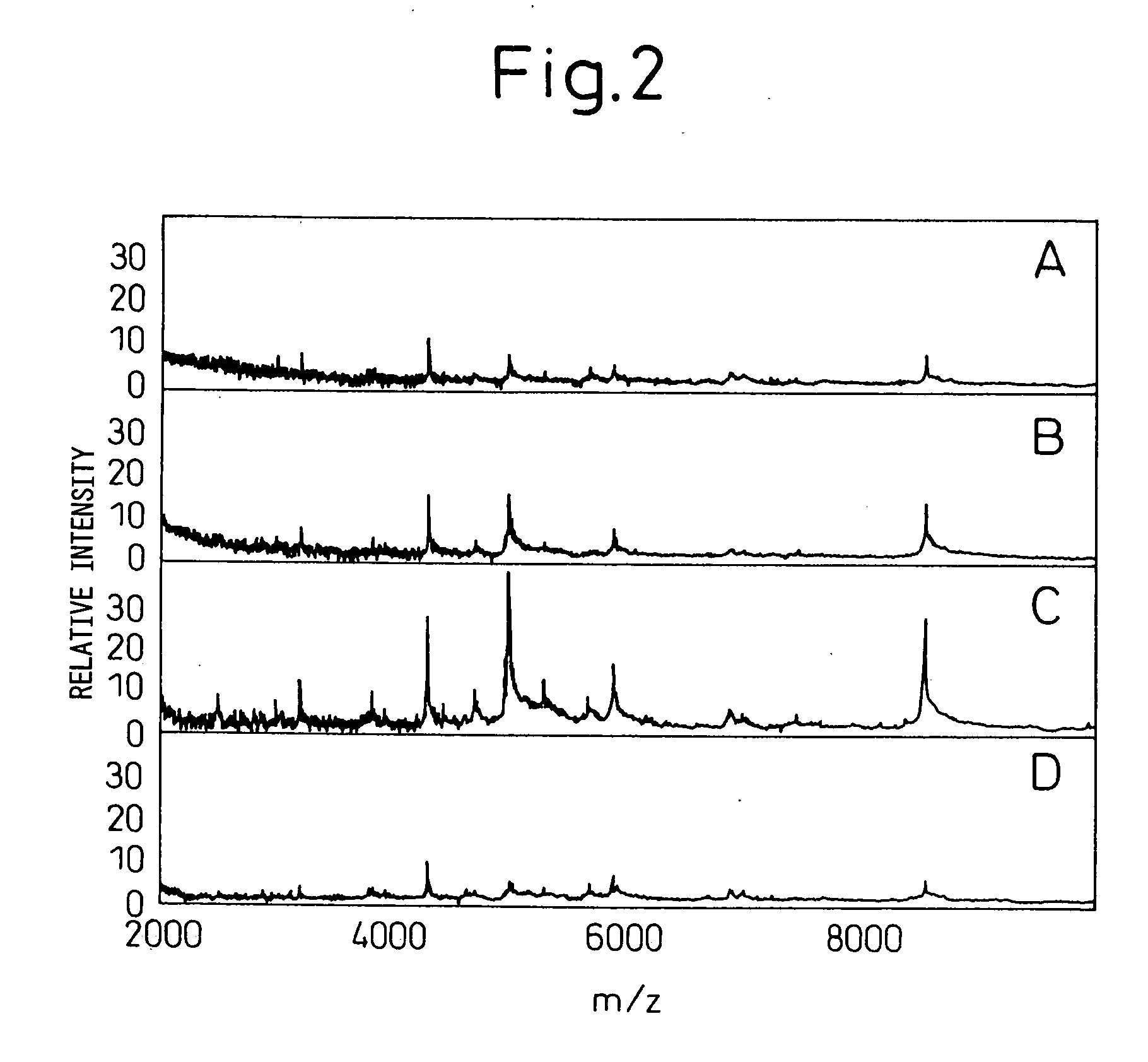
[0105] Using the protocol in Example 2, low molecular weight molecules were analyzed for five cell lines using 5 μl each. Protein concentration varied depending on the cell line (0.05-0.8 mg / ml), being equivalent to 250 ng to 2 μg analyzed per spot as the amount of total protein. Data were compared with those obtained by tricine SDS-PAGE that has been established as a technique for protein separation [Schagger et al., Anal. Biochem. 166:368-379 (1987)]. In order to perform tricine SDS polyacrylamide gel electrophoresis, 100 μl of the culture supernatant was concentrated to 20 μl, which was then dissolved in the sample buffer.
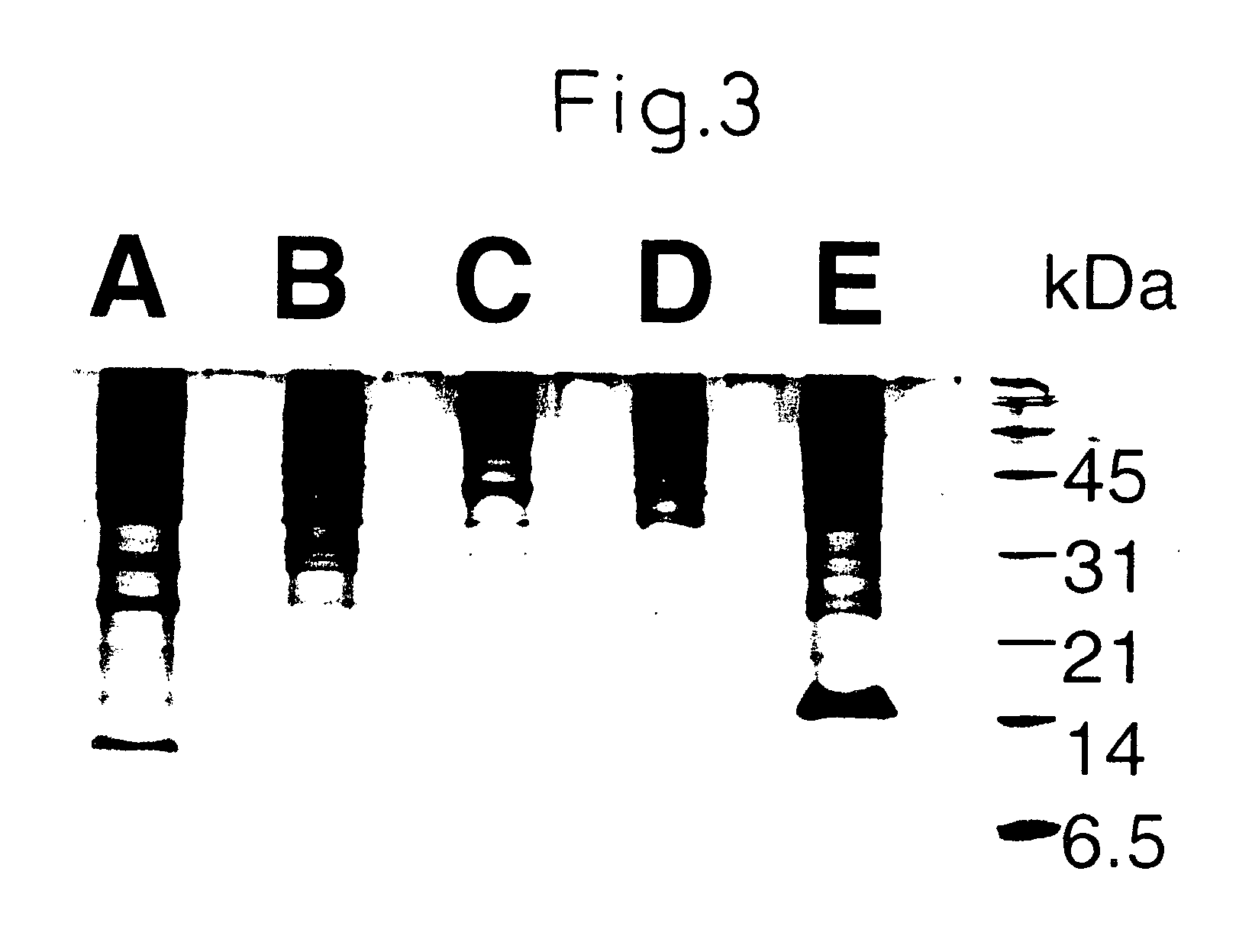
[0106] The gel composition used was 16.5% T-3% C. The bands were visualized by silver staining. Even when 100 μl of sample was used, electrophoresis detected only a small number of bands in the range of molecular weight 20000 or lower (FIG. 3). In the SELDI analysis, on the other hand, many signals were f...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| v/v | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com