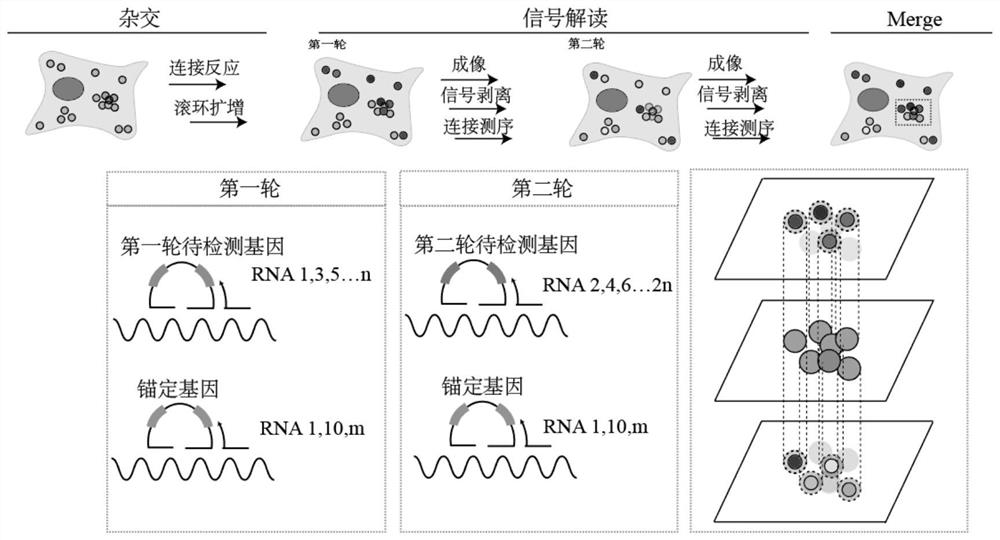
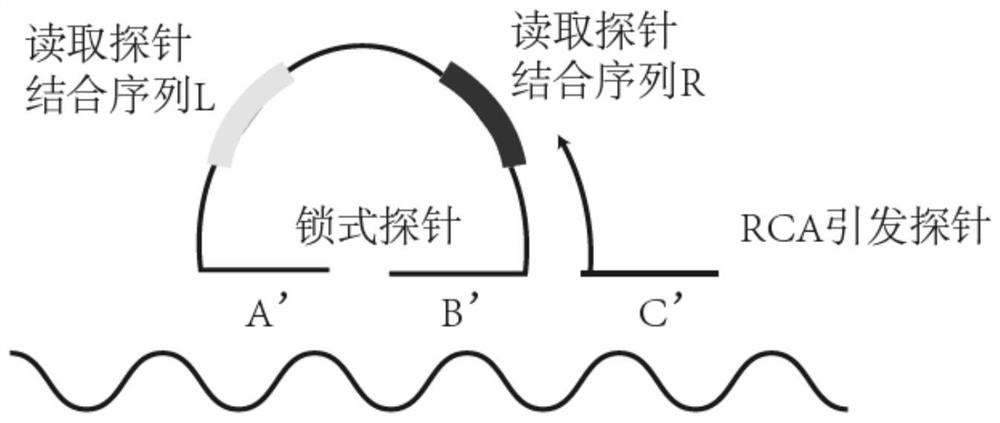
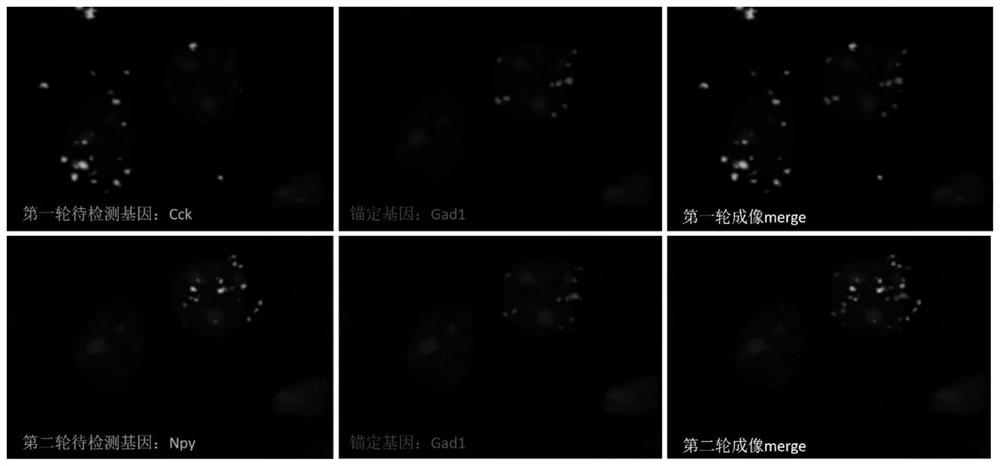
High-density signal decomposition sparse interpretation method applied to space omics and application of high-density signal decomposition sparse interpretation method
A space omics and signal decomposition technology, applied in the field of high-throughput in situ sequencing, can solve the problems of limited detection throughput, complicated operation steps, cumbersome fluorescent reading probe cost, etc., and achieve simple operation and difficult signal registration Low, the effect of reducing the difficulty of registration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Interpretation of 3 highly expressed genes in the cortex by using the method of spatial omics high-density signal decomposition and sparse interpretation: including the following steps:
[0050] 1. Selection of genes and design of hybridization probes
[0051] The expression gene Gad1 in the mouse cerebral cortex was selected as the anchor gene, and the highly expressed genes Cck and Npy were selected as the genes to be detected, and were assigned different barcodes respectively. The probe sequences were as follows:
[0052]
[0053]
[0054] 2. Probe Pretreatment
[0055] Apply T4 polynucleotide kinase to phosphorylate the 5' end of the padlock probe;
[0056] 3. Tissue sample collection
[0057] The brains of the mice sacrificed by cardiac perfusion were taken out, fixed in 4% PFA at 4°C for 12 hours, then dehydrated in 30% sucrose for 12 hours, and embedded in OCT for later use; the embedded mouse brains were sliced with a cryostat, The thickness is 10 μm,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com