Streptococcus suis efficient transposition mutation system and application thereof
A high-efficiency technology for Streptococcus suis, which is applied in the field of high-efficiency transposition mutation system for Streptococcus suis, and can solve the problems of not finding a high-efficiency random transposition mutation system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
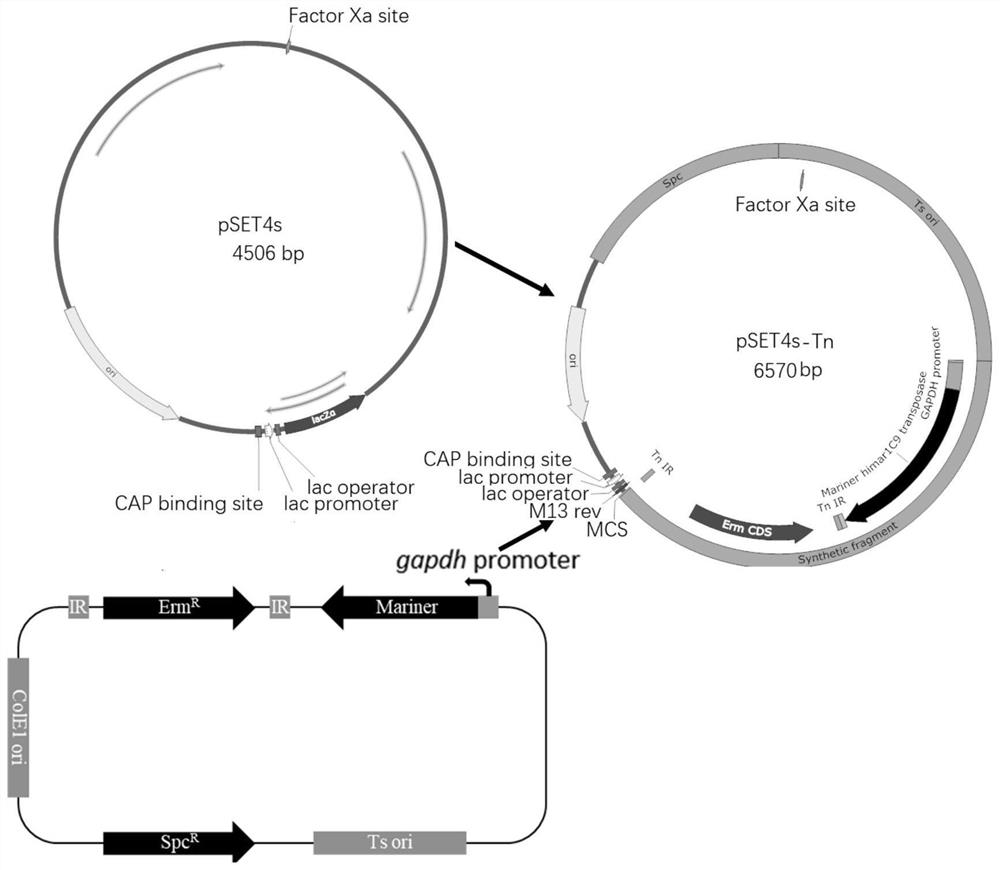
[0027] The Streptococcus suis gene inducible expression system of the present invention comprises a Mariner transposase expression cassette composed of a gapdh gene promoter and a transposase gene Mariner, and an erythromycin resistance expression cassette (Erm R ) connection composition (as shown in SEQ ID NO.2), named as Tn fragment, the nucleotide sequence of gapdh gene promoter is as shown in SEQ ID NO.14, the nucleotide sequence of transposase gene Mariner is as shown in SEQ ID NO. .15, the erythromycin resistance expression cassette (Erm R ) nucleotide sequence shown in SEQ ID NO.18, erythromycin resistance expression cassette (Erm R ) including the erythromycin resistance gene, the transposase recognition sequence (shown in SEQ ID NO. 17) between the erythromycin resistance gene and the Mariner transposase expression cassette, and the transposase recognition sequence on the other side (as shown in SEQ ID NO. 16).
[0028] The Streptococcus suis high-efficiency transpo...
Embodiment 2
[0057] Example 2 Construction of Streptococcus suis transposition mutants using pSET4s-Tn plasmid
[0058] (1) Add about 100 ng of pSET4s-Tn plasmid to the competent Streptococcus suis, mix well on ice, and perform click transformation, then add 400 μl of TSB to culture at 28°C for 2 hours, and then spread the culture with the final concentration 5% bovine serum, 10 μg / mL erythromycin and 100 μg / mL spectinomycin on TSA culture plate, incubator at 28℃ for overnight culture;
[0059] (2) Using the primer pair shown in SEQ ID NO. 6 and SEQ ID NO. 7 and the primer pair shown in SEQ ID NO. 8 and SEQ ID NO. 9: Erm-F: 5'-GAAACCGATACCGTTTACG-3' Colony PCR with Erm-R: 5'-GACGATATTCTCGATTGACC-3' and Spc-F: 5'-GATCAGGAGTTGAGAGTG-3' and Spc-R: 5'-CTCTTCTCACATCAGAAAATGG-3' to confirm the successful transformation of the plasmid into S. suis;
[0060] (3) Inoculate a single colony of Streptococcus suis containing pSET4s-Tn plasmid into a TSB liquid medium containing a final concentration o...
Embodiment 3
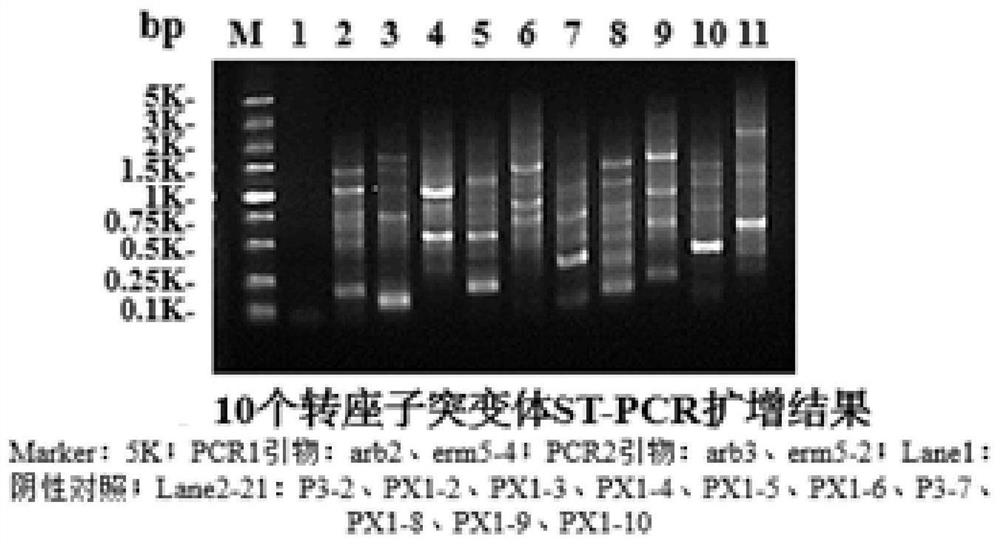
[0063] Example 3 Identification of transposon insertion sites
[0064] (1) 10 Streptococcus suis transposon mutants were randomly selected, inoculated into TSB liquid medium containing a final concentration of 5% bovine serum and 5 μg / mL erythromycin, and cultured at 37°C for 12 hours, using genomic DNA Extraction kits extract bacterial genomes.
[0065] (2) Using the genomic DNA extracted above as a template, using primers arb-2: 5'-GGCCACGCGTCGACTAGTCANNNNNNNNNNGATCA-3' and erm5-4: 5'-GGTTGAGTACTTTTTCACTCG- 3' for the first round of PCR. The reaction system is: upstream and downstream primers 0.5nM each, genomic DNA 2μL, PCR Mix 10μL, ddH 2 O supplemented to 20 μL; PCR reaction conditions were 94°C, 2 min; 94°C, 30s; 42°C, 30s; 72°C, 3min, the above reaction was repeated for 6 cycles; 94°C, 30s; 65°C, 30s; 72°C, 3min , the above reaction was repeated for 25 cycles; 4°C, 10min;
[0066] (3) Using one round of PCR products as templates, primers arb-3 as shown in SEQ ID NO....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com