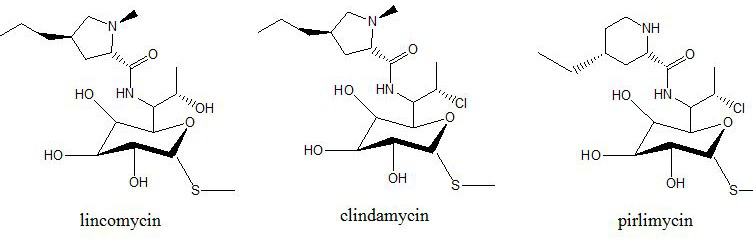
Method for determining lincosamine antibiotics in feed
A technology for antibiotics and lincomycin, applied in the field of determination of lincomycin antibiotics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
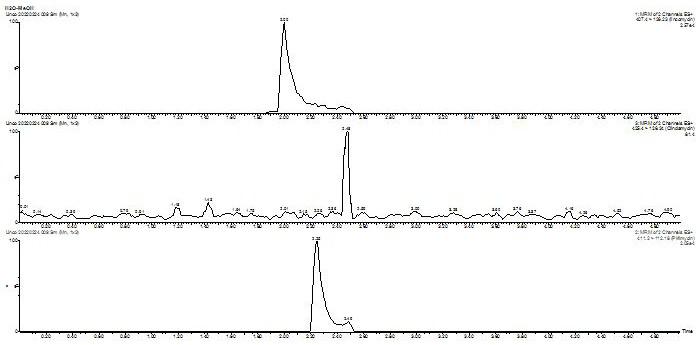
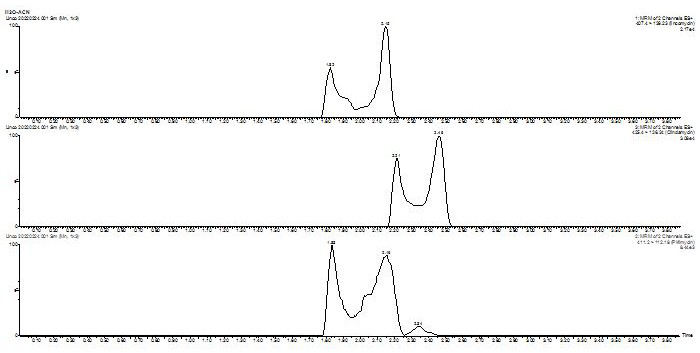
[0032] Example 1 LC-MS / MS analysis conditions
[0033] The mobile phase consists of two phases A and B (A: 0.1% formic acid in water; B: acetonitrile), gradient elution, see Table 1 for the elution procedure; the flow rate is 0.4 mL min -1 ; the column temperature was set to 30°C; the injection volume was 8 µL.
[0034]
[0035]Lincomycin, clindamycin, and piromycin all have tertiary amine structures and are weakly basic. They have a strong positive ion response in mass spectrometry. Therefore, positive ion mode scanning and multi-stage reaction monitoring ( MRM). The atomizing gas and drying gas are high-purity nitrogen, and the collision gas is high-purity argon. Prepare a concentration of 100 μg L with A and B mobile phases (50:50, V / V) -1 3 compounds lincomycin, clindamycin, and piromycin, 10 mL min by syringe pump -1 The flow rate was injected into mass spectrometry to optimize mass spectrometry parameters such as cone voltage, collision energy, dwell time, and fra...
Embodiment 2
[0037] Example 2 Methodological study
[0038] The method of the present invention was validated with reference to European Commission Decision No. 2002 / 657 / EC (2002 / 657 / EC) to evaluate matrix effects, accuracy (recovery), precision (reproducibility), linearity and selectivity, and Minor modifications to it: Limit of detection (LOD) and limit of quantification (LOQ) are used instead of limit of detection determination (CCα) and detection capacity (CCβ) commonly found in animal feed (Frenich et al., 2011; Ying et al., 2013; Kiebooms et al, 2015)
[0039] In the present invention, the matrix effect (ME) is assessed as the ratio of the slope of the working curve of the matrix-matched standard solution to the slope of the working curve of the standard solution of pure solvent (Chawla et al., 2017; Varga et al., 2021). For the preparation of a series of mixed standard working solutions (1.0, 2.0, 5.0, 10.0, 50.0, 100.0 and 200 μg L -1 ). The matrix effect was calculated accordin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com