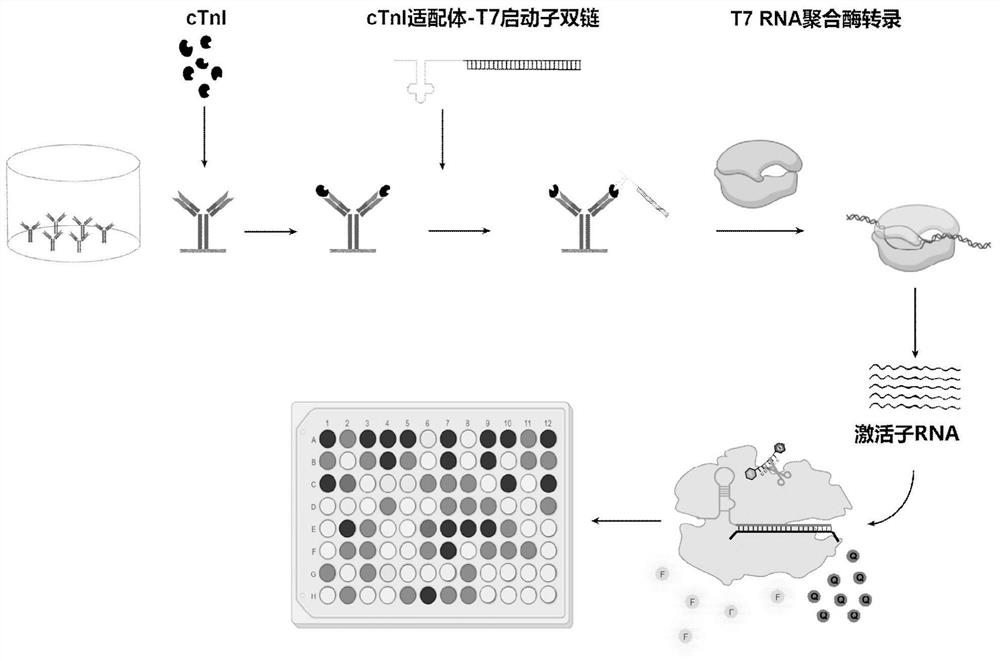
Method for detecting cardiac troponin I based on CRISPR Cas13d
A technique for cardiac troponin and analyte, applied in the field of fluorescence detection, can solve the problem of detecting non-nucleic acid molecules without Cas13d, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
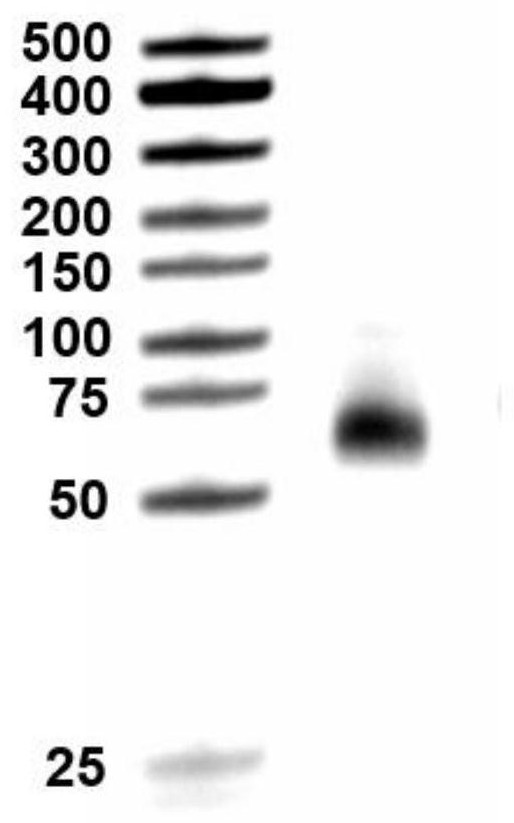
[0056] CRISPR RspCas13d protein expression and purification, the specific operation includes the following steps:
[0057] 1. 3 μl of pET28a-MH6-RspCas13d plasmid and E. coli DE3 competent cell BL21 (50 μl) were placed in an ice bath and incubated in a clean bench for 10 min;
[0058] 2. After the competent cells were heat-shocked in a 42°C water bath for 90s, they were immediately taken out and placed in an ice bath to cool for 5min;
[0059] 3. Add 450 μL of anti-antibody LB liquid medium to the competent cells introduced into the plasmid in step 2, invert and mix, and incubate at 37°C with shaking at 220 rpm for 1 hour;
[0060] 4. Centrifuge and aspirate 400 μL of the supernatant of the medium in step 3 and discard it, and resuspend the remaining bacterial liquid, spread it evenly on the AMP-resistant LB solid plate medium, and cultivate it upside down at 37 degrees for 12 hours;
[0061] 5. Pick a single clone into 5ml of AMP-resistant LB liquid medium and culture at 37°...
Embodiment 2
[0069] CRISPR RNA synthesis, the specific operation includes the following steps:
[0070] The CRISPR RNA required for the present invention was obtained by HyperScribe TM T7 High Yield RNA SynthesisKit (Apexbio) was synthesized by in vitro transcription.
[0071]
[0072] The reagents required for the synthesis process are shown in the table above, and the main steps are as follows:
[0073] 1. Prepare crRNA-F and crRNA-R DNA primers in DEPC water (see the primer sequence list) and determine the concentration to 10 μM in a micro UV spectrophotometer;
[0074] 2. The above ssDNA was annealed in annealing buffer (20 mM Tris-HCl, pH=8.0, 100 mM NaCl, 8 mM MgCl at a ratio of 1:1) 2 ) annealed and assembled into double-stranded DNA;
[0075] 3. The DNA double-stranded DNA in step 2 is mixed according to the added amount of each reagent in the table above, and then mixed with the pipette tip and incubated at 37 degrees for 16 hours;
[0076] 4. Add 3 μL of DNase I 10× react...
Embodiment 3
[0079] CRISPR RNA purification by spin column method, the specific operation includes the following steps:
[0080] The RNA purification kit used in this example is Monarch RNA Cleanup Kit T2040 (50 μg) (NewEngland Biolabs). RNA is bound to the silica adsorption column by a high-concentration salt ion binding buffer, and washed with a low-salt ion elution buffer. The impurities bound to the silica column are removed, so as to achieve the purpose of RNA purification.
[0081] Refer to the manufacturer's instructions for the specific operation process. The main process is as follows:
[0082] 1. Add 20 μL of RNase-free double-distilled water to the Mix obtained in step 5 in Example 2 to a volume of 50 μL;
[0083] 2. Add 100 μL RNACleanup binding buffer to Mix in step 1;
[0084] 3. Add 150μL of ethanol (purity > 95%), mix by pipetting with pipette tip;
[0085] 4. Transfer all the mixture in 3 to the spin column, centrifuge at 12000rpm for 30s, and discard the flow-through l...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com