Process for producing peptide
A technology of target peptide and cystyl peptide, applied in the field of peptide preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
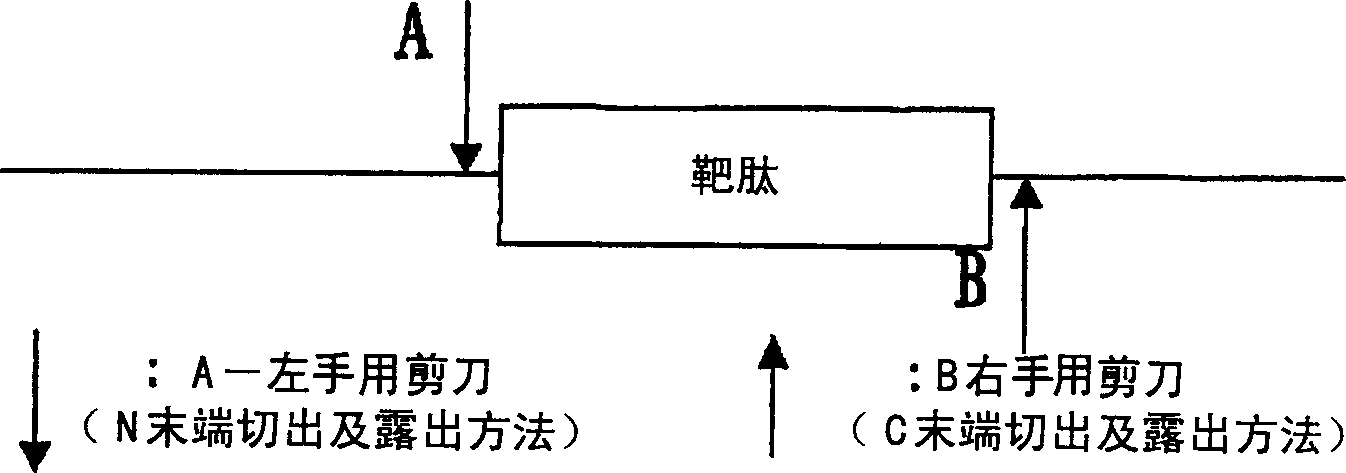
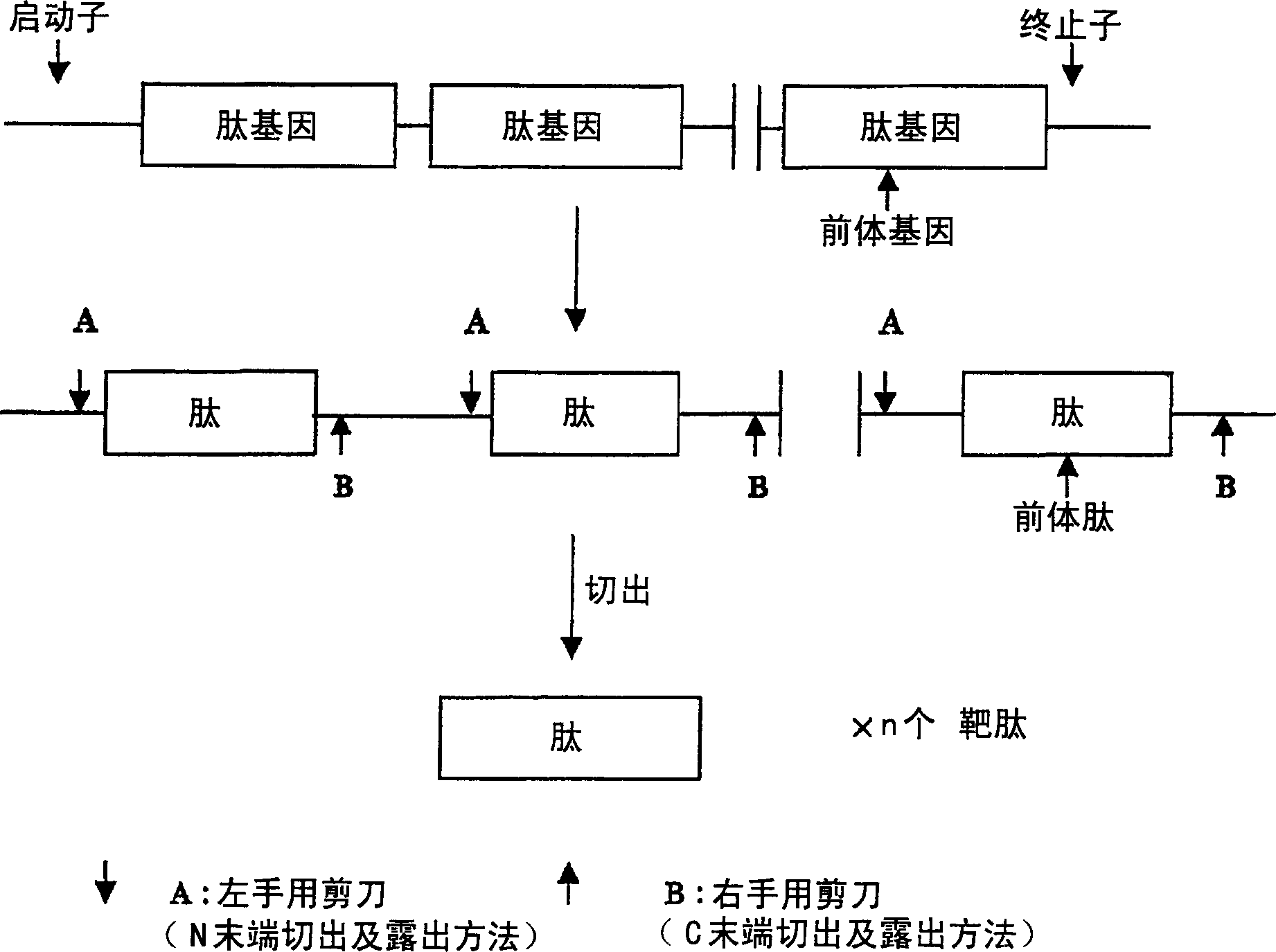
[0076] In the production method A of the present invention, as the cleavage site of the enzyme that can be added to the N-terminal of the target peptide, such as
[0077] (1) Asp-Asp-Asp-Asp-Lys (SEQ ID NO: 58) (encoded by DNA containing base sequence: GATGACGACGACAAG (SEQ ID NO: 59)) which is a cleavage site of proteolytic enzyme enterokinase, etc.
[0078] (2) Ile-Glu-Gly-Arg (SEQ ID NO: 60) (encoded by DNA containing base sequence: ATTGAAGGCCGC (SEQ ID NO: 61)) which is a cleavage site of proteolytic enzyme factor Xa, etc.
[0079] (3) Gly-Pro-Arg (SEQ ID NO: 62) (encoded by DNA containing the base sequence: GGCCCGCGC (SEQ ID NO: 63)) which is the cleavage site of the proteolytic enzyme thrombin, etc.
[0080] As a chemical cleavage site that can be added to the N-terminal of the target peptide, for example, a methionine residue as a cyanogen bromide cleavage site, etc.
[0081] As a chemical cleavage site that can be added to the C-terminus of the target peptide, such as ...
Embodiment 1
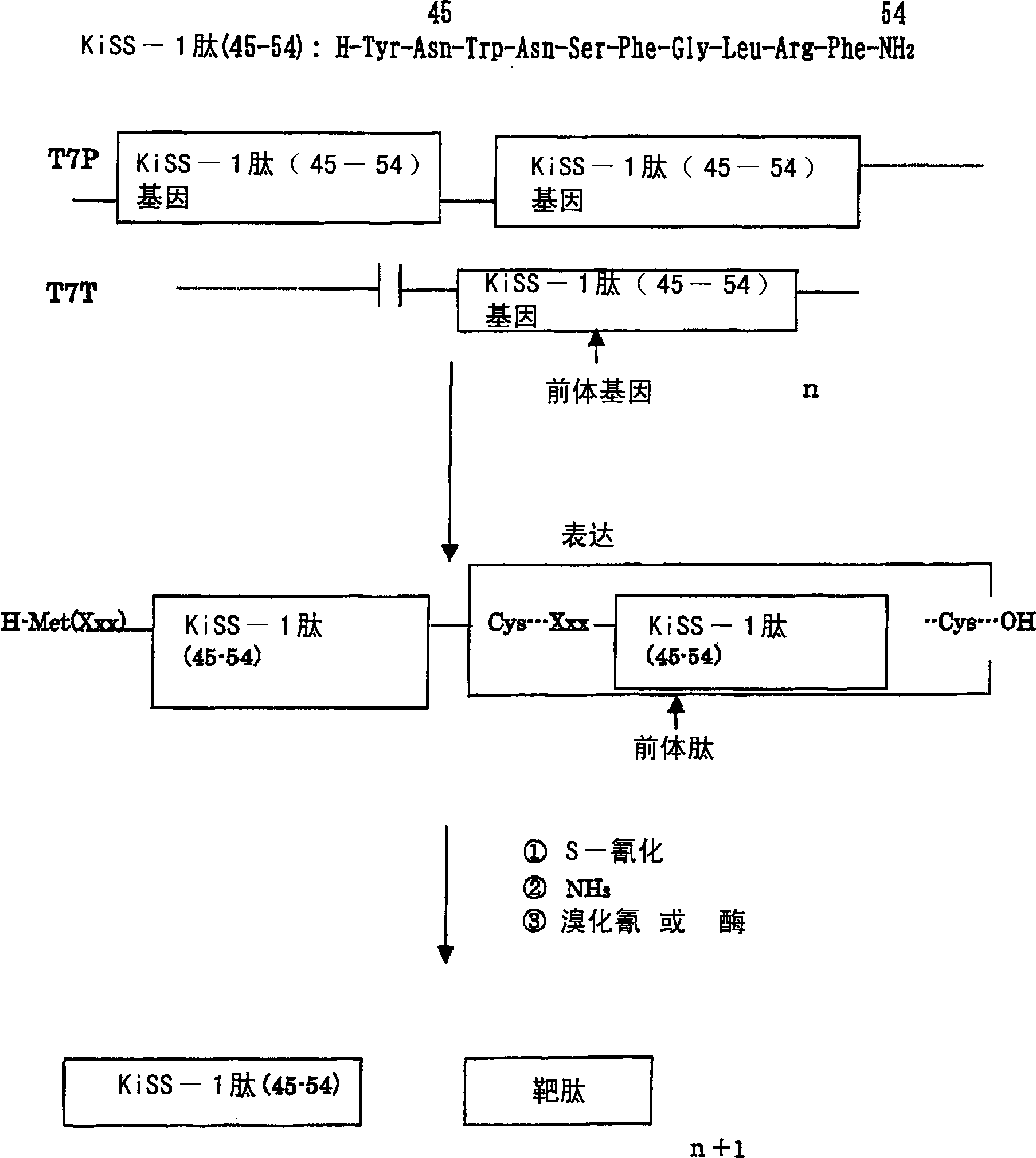
[0263] (a) Preparation of gene encoding human GPR8 ligand (hGPR8L; SEQ ID NO: 44) in tandem three times
[0264] The following 10 kinds of DNA fragments (sequences 65-74 in the sequence listing) were used to prepare a structural gene encoding human hGPR8L ligand in tandem three times.
[0265] #1
[0266] 5'-
[0267] TATGGATGACGATGACAAATGGTAAAAACATGTGGCGAGCCCGCGTT
[0268] ATCATA CCG
[0269] (Sequence 65)
[0270]#2
[0271] 5'-
[0272] GCGCGGCCCACGGTATGATAACGCGGGCTCGCCACATGTTTATACCA
[0273] TTTGTCATCGTCATCCA
[0274] (Sequence 66)
[0275] #3
[0276] 5'-
[0277] TGGGCCGCGCGGCCGGTCTGCTGATGGGCCTGTGTCAATTGGGTTT
[0278] GAACTTCTCTGTCTCCGCCGCCGGAG
[0279] (Sequence 67)
[0280] #4
[0281] 5'
[0282] GATCCTCCGGCGGCGGAGACAGAGAAGTTCAAACCCAATTGACACA
[0283] GGCCCATCAGCAGACCGGCC
[0284] (Sequence 68)
[0285] #5
[0286] 5'-
[0287] AATTGGGTGGTGATGACGATGACAAATGGTATAAACATGTGGCGAGC
[0288] CCGCGTTATCATACCG
[0289] (Sequence 69)
[0290] #6
[0291] ...
Embodiment 2
[0322] Add 10 ml of 10 mM EDTA (pH6.0) to 2 g of the cells obtained in Example 1, and after ultrasonic treatment (BRANSON SONIFIER MODEL450), centrifuge (15000 rpm for 15 minutes). Repeat the same operation for the precipitate. After adding 5 ml of 7M guanidine solution (pH 5.0) to the precipitate, it was stirred for 2 hours and then centrifuged (15000 rpm for 15 minutes). Add 17 mg of Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP-HCl) to the supernatant, perform a reduction treatment at 50° C. for 10 minutes, and then pass it through C4P-50 (1 cm×25 cm, Showa Electrician), after adsorption and cleaning, carry out gradient elution of 20-60% B (B: 80% acetonitrile / 0.1% trifluoroacetic acid) at a flow rate of 2ml / min, and collect the precursor protein fraction (elution time approx. 27 minutes), freeze-dried to obtain the freeze-dried powder of the precursor protein.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com