Single molecule dsDNA microarray chip preparation method
A microarray chip and microarray technology, applied in biochemical equipment and methods, microbe measurement/inspection, etc., can solve the problems of reducing the number of ideal double-stranded nucleic acid probes, the existence of probe stability, and high cost of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
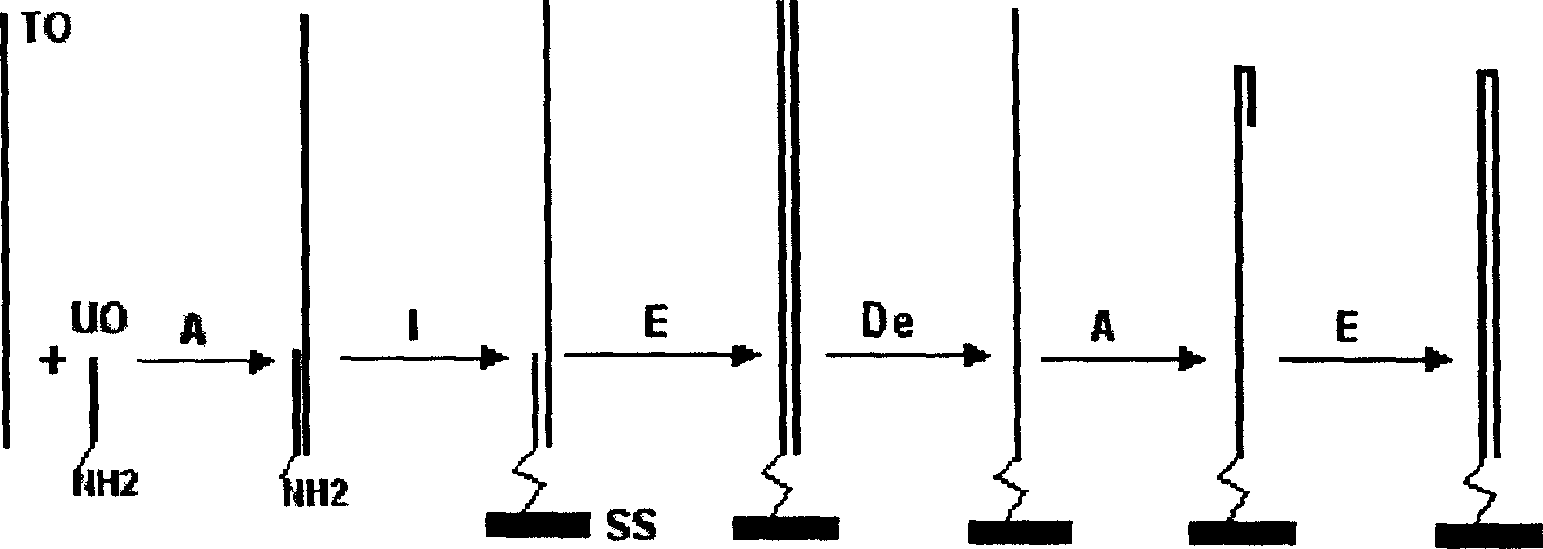
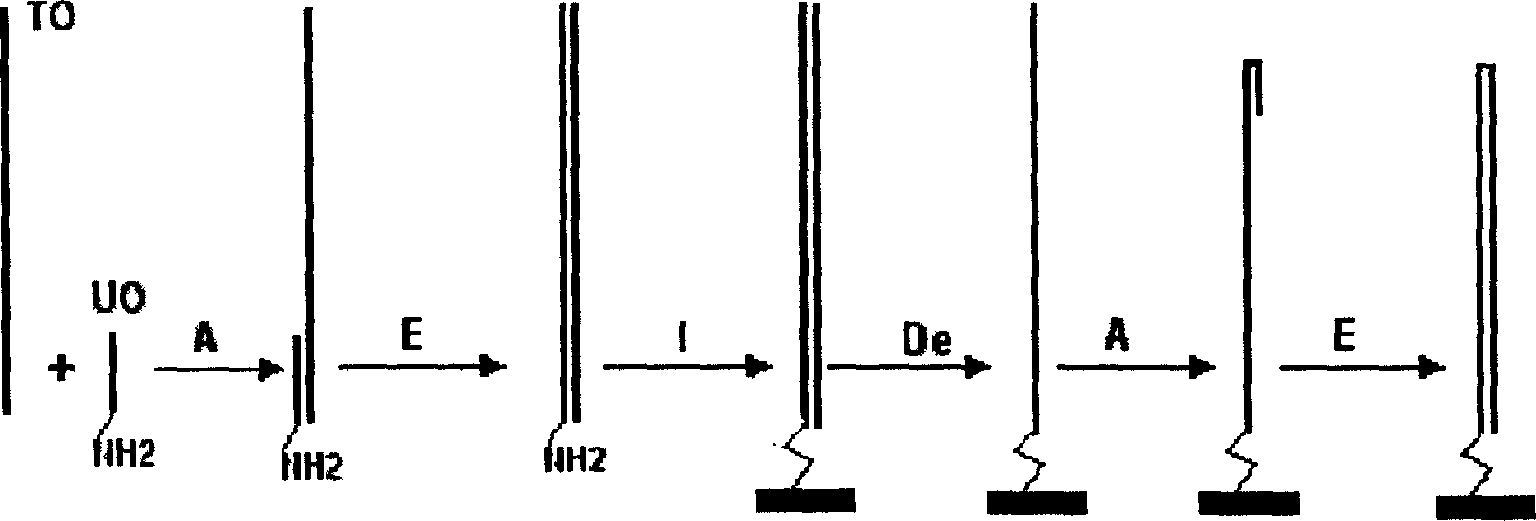
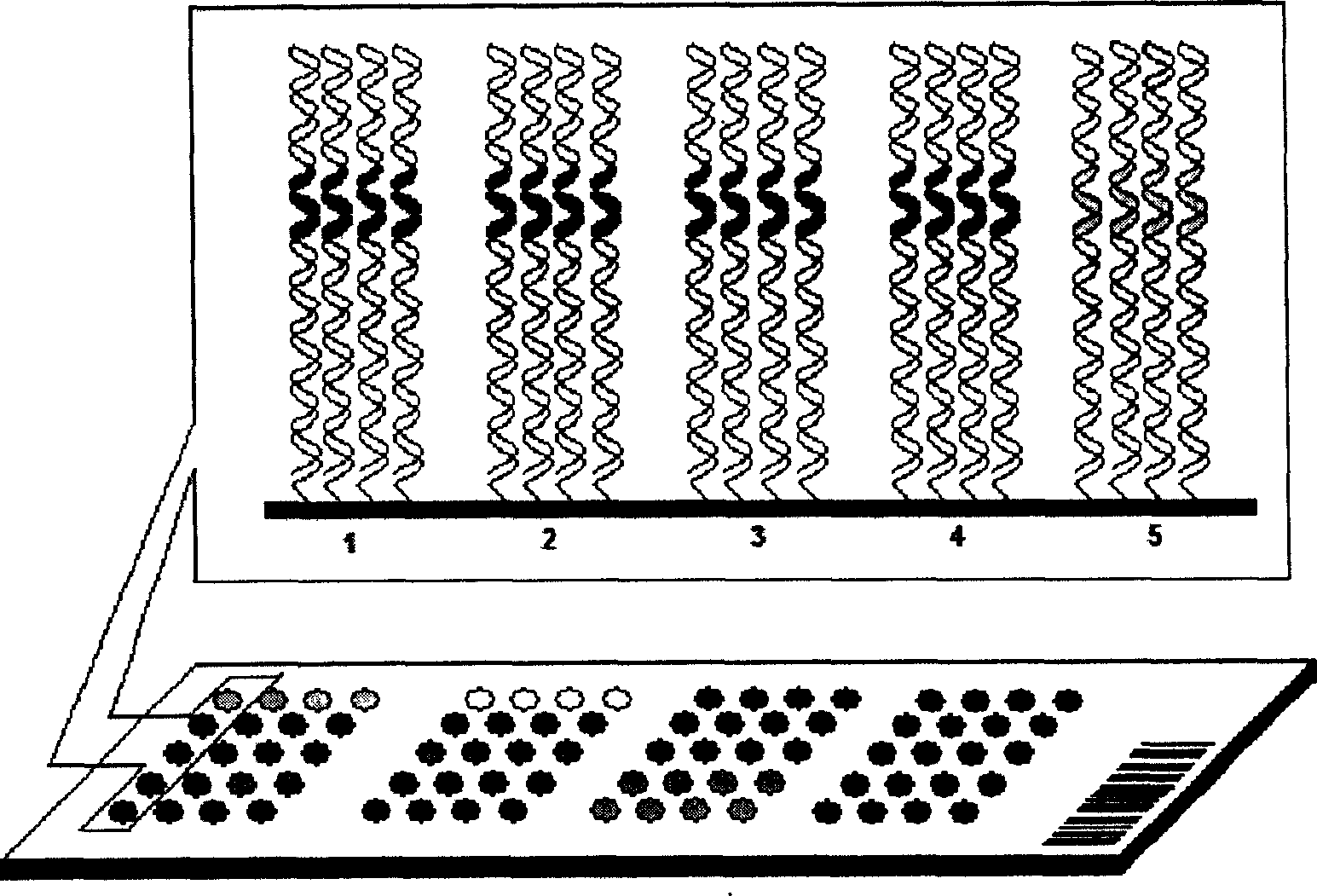
[0080] Here, the single-molecule dsDNA microarray chip containing the same DNA probe for detecting NF-κB protein is prepared on the chip as an example by using the glass slide treated with aminosilane on the surface as a solid phase support to illustrate the single-molecule dsDNA microarray chip of the present invention. Specific implementation of array chip preparation and application.
[0081] 1. Slide preparation, silanization and aldehyde modification:
[0082] If commercially available slides for gene chips such as SuperAldehyde Slides from Telechem are used, this step does not need to be performed; if the slides are prepared from ordinary tissue sections, the slides are not allowed to be processed. Firstly, the slides were routinely cleaned, and then silanized with a silylating agent such as aminopropyltriethoxysilane (triethoxyaminosilane) from Sigma for 5-10 minutes (95% acetone containing 2% aminopropyltriethoxysilane, acetone). After silanization, wash twice with de...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com