Human calcium ion / calmodulin deopendent protein kinase II inhibitory protein alpha, its coded sequence and use
A technology of sequence and coding, applied in the fields of biotechnology and medicine, which can solve the problem of non-discovery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] Cloning of Human CaM-KIINα cDNA
[0098] Total RNA of human dendritic cells was extracted with Trizol reagent (Life Technologies). Then, poly(A) mRNA was isolated from total RNA. After the poly(A) mRNA was reverse-transcribed to form cDNA, the cDNA fragment was directional inserted into the multiple cloning site of the vector using the SuperScript II cloning kit (purchased from Gibco), and transformed into DH5α bacteria to form a cDNA plasmid library. The 5' termini of randomly selected clones were sequenced by the dideoxy method. Comparing the determined cDNA sequence with the existing public DNA sequence database, it was found that the DNA sequence of one cDNA clone was a new full-length cDNA. The DNA sequence contained in the new clone is bidirectionally determined by synthesizing a series of primers. Computer analysis showed that the full-length cDNA contained in the clone was a new cDNA sequence (shown in SEQ ID NO: 1) encoding a new protein (shown in SEQ ID NO:...
Embodiment 2
[0102] Obtaining the Coding Sequence of Human CaM-KIINα by RT-PCR and Analysis of Its Expression in Cells
[0103] (1) Obtain human CaM-KIINα coding sequence by RT-PCR method
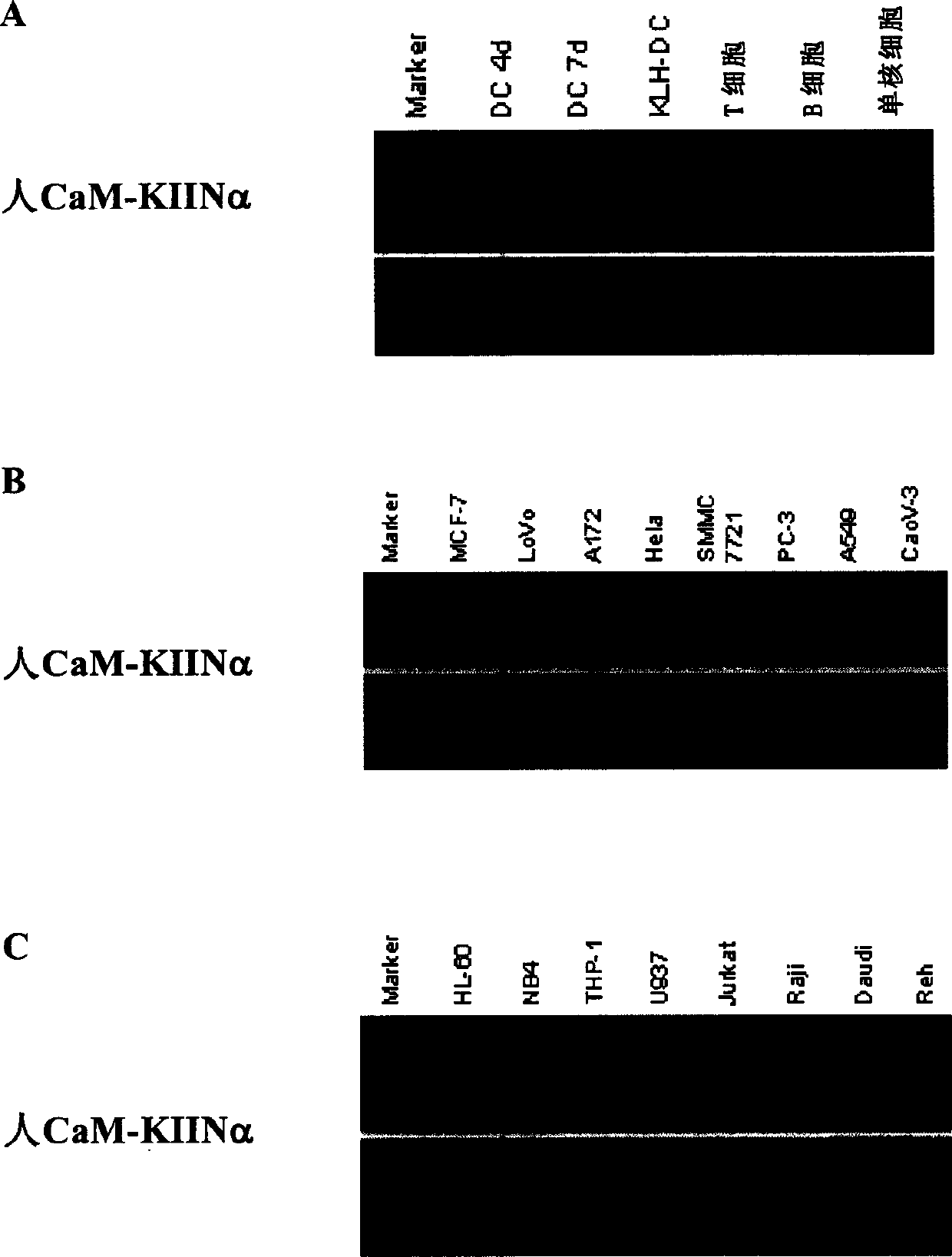
[0104]The total RNA of dendritic cells derived from corresponding cell lines in the logarithmic growth phase, human peripheral blood mononuclear cells and human peripheral blood mononuclear cells stimulated by LPS for different times was extracted with Trizol reagent, and 5 μg of total cellular RNA was mixed with 1 μg Oligo- dT12-18 were mixed for reverse transcription. The reverse transcription system is 20 μl, and 80 μl ddH2O is added after the reaction for dilution. The primers used for PCR amplification of CaM-KIINα are as follows: sense primer 5' CCG AGG ATG TGA GTCCTG CTC (SEQ ID NO: 3), antisense primer 5' GAG GAG CCC AGA GCC TTC TC (SEQ ID NO: 4), At the same time, β-actin was used as a positive control. The PCR reaction volume was 50 μl, which contained 10 μl of reverse transcription templat...
Embodiment 3
[0109] Recombinant expression of human CaM-KIINα
[0110] In this example, using the RT-PCR amplification product obtained in Example 2 as a template, PCR oligonucleotide primers with the following sequences at the 5' and 3' ends were used to amplify to obtain the full-length code of human CaM-KIINα region DNA as the insert.
[0111] The 5' end oligonucleotide primer sequence used in the PCR reaction is:
[0112] 5'-AC GGA TCC ATG TCG GAG GTG CTG CCC T-3' (SEQ ID NO: 5)
[0113] The primer contains an enzyme cutting site of BamH I restriction endonuclease, behind the enzyme cutting site is the entire coding sequence of human CaM-KIINα;
[0114] The 3' end primer sequence is:
[0115] 5'-TC GAA TTC TTA GAC ACC AGG AGG TGC CT-3' (SEQ ID NO: 6)
[0116] The primer contains the cutting site of EcoR I restriction endonuclease, the translation terminator and the entire coding sequence of human CaM-KIINα.
[0117] Using the RT-PCR amplification product obtained in Example 2 as a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com