I type aldehyde dehydrogenase of clonorchis sinensis, its coding nucleic acid and application
A technology of Clonorchis sinensis and aldehyde dehydrogenase, which can be used in application, oxidoreductase, anti-enzyme immunoglobulin, etc., and can solve problems such as low expression level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment I
[0097] Example I: Cloning of Clonorchis sinensis Class I Aldehyde Dehydrogenase
[0098] The total RNA of Clonorchis sinensis adults was extracted by one-step method of guanidine isothiocyanate / phenol / chloroform. Poly(A) mRNA was isolated from total RNA using Quik mRNA Isolation Kit (product of Qiegene). 2ug poly(A) mRNA was reverse transcribed to form cDNA. Smart cDNA cloning kit (purchased from Clontech) was used to insert the cDNA fragment into the multiple cloning site of the pBSK(+) vector (product of Clontech Company), transform DH5α, and the bacteria formed a cDNA library. The sequences of the 5' and 3' ends of all clones were determined with Dye terminate cycle reactions sequencing kit (product of Perkin-Elmer) and ABI 377 automatic sequencer (Perkin-Elmer). Comparing the determined cDNA sequence with the existing public DNA sequence database (Genebank), it was found that the cDNA sequence of one of the clones, c002D06, was a new DNA. The insert cDNA fragment contai...
Embodiment 2
[0099] Example 2: Homology retrieval of cDNA clones
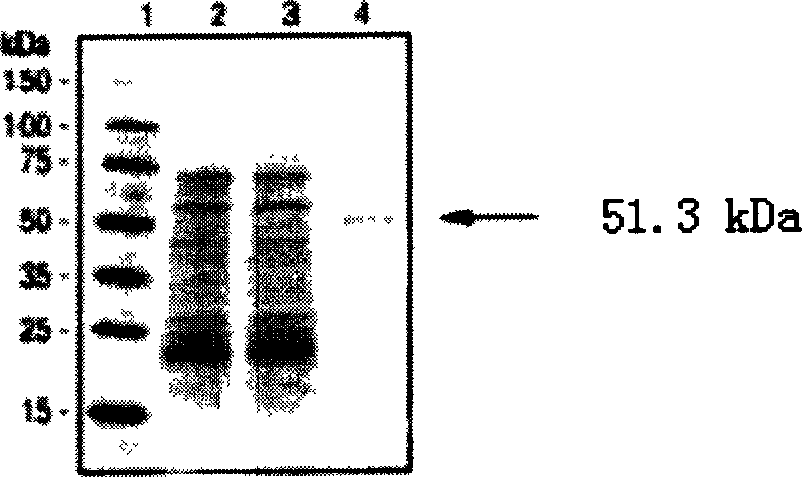
[0100] With the sequence of the Clonorchis sinensis class I aldehyde dehydrogenase of the present invention and the protein sequence encoded thereof, use the Blast program (Basiclocal Alignment search tool) [Altschul, SF et al.J.Mol.Biol.1990; 215:403 -10], homologous searches were performed in databases such as Genbank and Swissport. The gene with the highest homology to the Clonorchis sinensis class I aldehyde dehydrogenase of the present invention is a known Xenopus class I aldehyde dehydrogenase, the two are highly homologous, and the protein homology results are shown in figure 1 , whose identity is 59%; similarity is 75%.
Embodiment 3
[0101] Embodiment 3: Cloning the gene encoding Clonorchis sinensis class I aldehyde dehydrogenase by RT-PCR method
[0102] The total RNA of Clonorchis adultes was used as template, and oligo-dT was used as primer to carry out reverse transcription reaction to synthesize cDNA. After purification with Qiagene kit, PCR amplification was carried out with the following primers:
[0103] Primer1: 5'-CTGTTCATTAATAATGAATTTGTG-3' (SEQ ID NO: 3)
[0104] Primer2: 5'-TCTTTTTTTTTTTTTTTTGGAGAATG-3' (SEQ ID NO: 4)
[0105] Primer1 is the forward sequence starting from the 1st bp at the 5' end of SEQ ID NO:1;
[0106] Primer2 is the 3' reverse sequence in SEQID NO:1.
[0107] The conditions of the amplification reaction: 50mmol / L KCl, 10mmol / LTris-Cl, (pH8.5), 1.5mmol / LMgCl in a reaction volume of 50μl 2 , 200 μmol / L dNTP, 10 pmol primer, 1 U of Taq DNA polymerase (product of Clontech Company). On a PE9600 DNA thermal cycler (Perkin-Elmer), the reaction was performed for 25 cycles under...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com