Coding calicheamicin biosynthesis and micromonospora echinospora gene for same antagonism
A biosynthetic technology of Micromonospora aculeatus, applied in the field of isolating genes and their corresponding proteins, can solve problems such as lack of knowledge
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
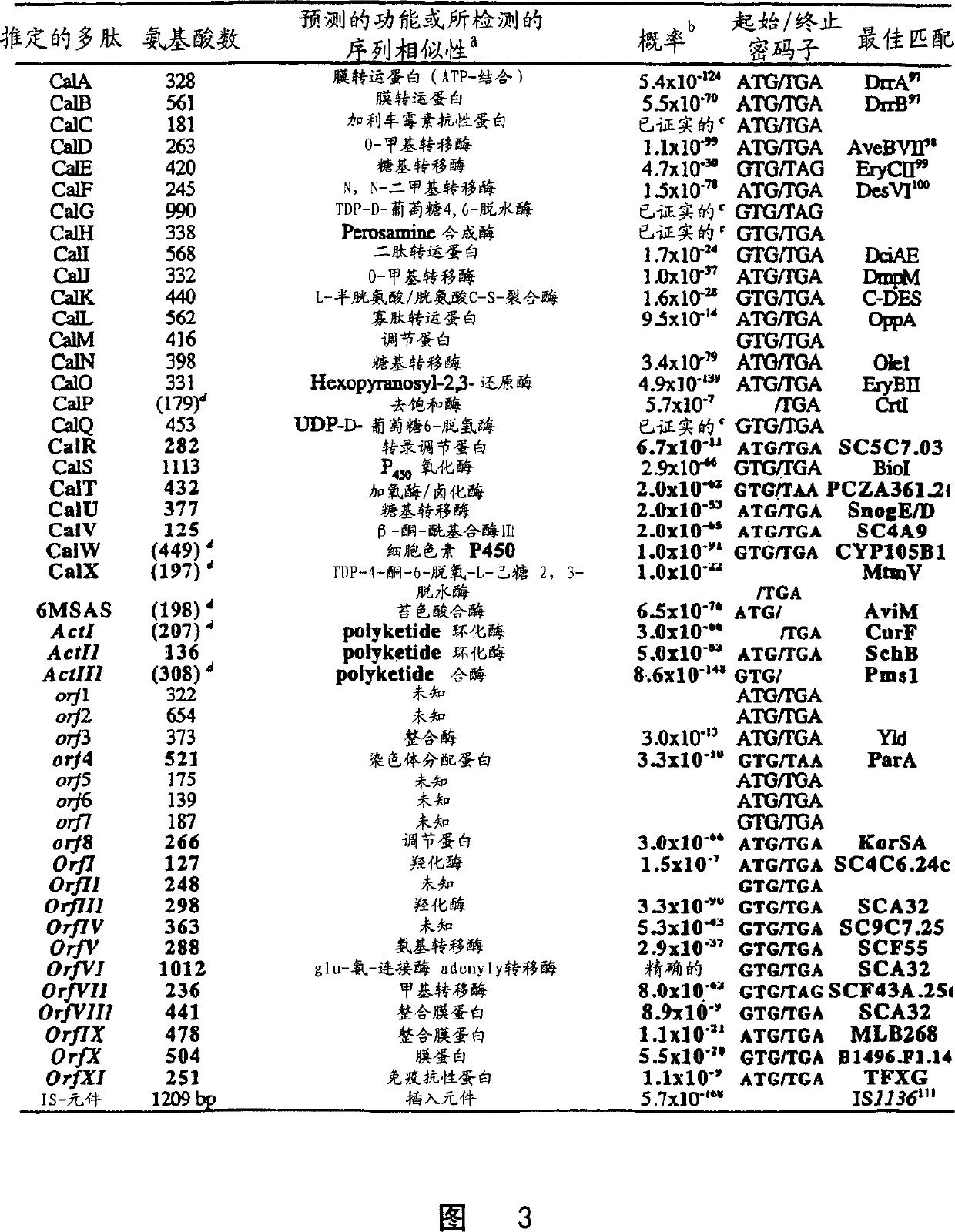
[0107] To elucidate nucleotide sequences as quickly as possible, thermal cycle sequencing was performed from pUC- or pBluescript-based subclones (using M13 primers and primer walking), but also directly from isolated cosmids (via primer walking). Nucleotide sequence data were obtained using two Applied Biosystems Auto 310 Genetic Analyzers, followed by Applied Biosystems AutoAssembler TM DNA sequence assembly software for assembly. Dear, S. et al., Nucl Acids Res., 14, 3907-3911 (1991); Huang, X., Genomics, 14, 18-25 (1992). The Orf arrangement was calculated using the calculation program MacVector TM Combination of 6.0 and Brujene. MacVector is a commercially available software package that has the ability to construct a Micromonospora codon bias table (from known Micromonospora sequences) and then use this codon bias table to retrieve optimal orfs. Fickett, J.W., Nucleis Acids Research, 10, 5303-5318 (1982). Alternatively, the trialware program Brujene is specially des...
Embodiment 2
[0108] Example 2: Isolation and characterization of calC
The maximum tolerated concentration of calicheamicin
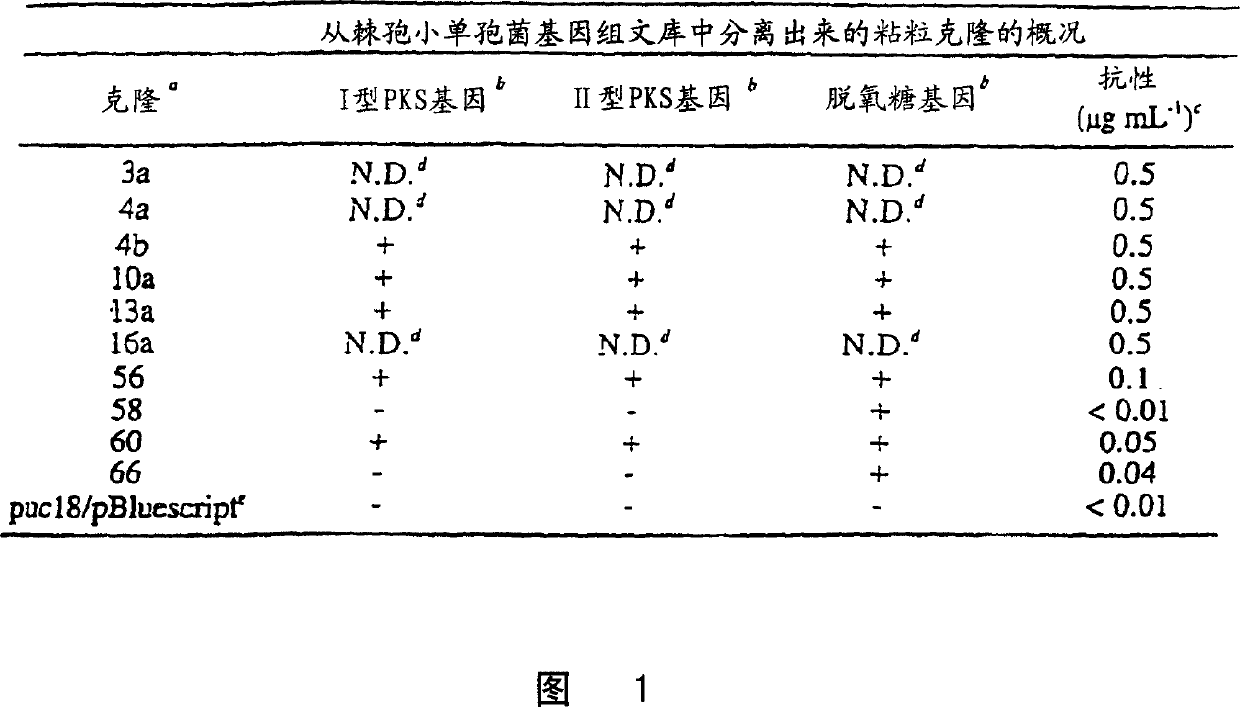
Cosmids 3a, 4a, 10a, 13a and 16a
0.5 μg ml -1
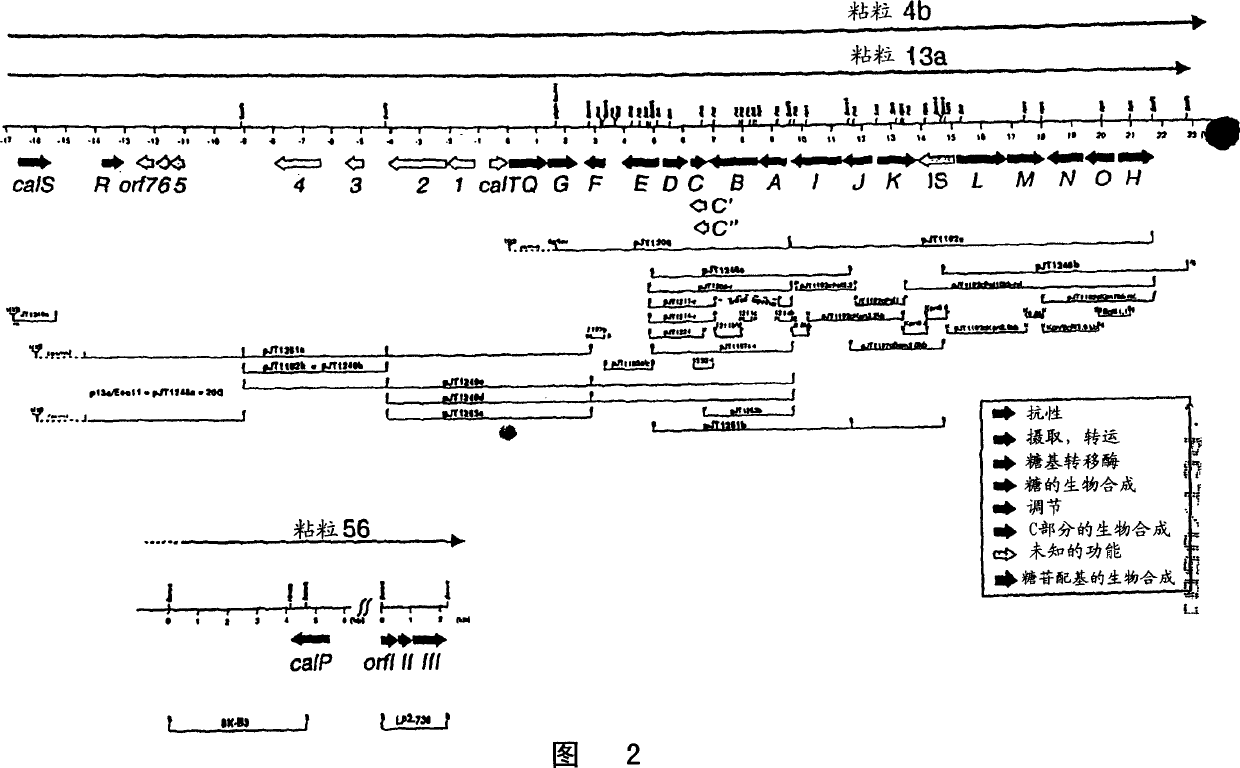
pJT1214 to pJT1232
5.0 μg ml -1
PRE7
20.0 μg ml -1
pRE7
50.0 μg ml -1
pJT1224, pAP6, Pre1, and control plasmids
pUC18, pBluescript, and pMALC2
-1
[0110] Nucleotide sequence analysis of the PstI-SacI fragment revealed that it contains two possible orfs. The proximal 1 kb of the fragment carries a single orf calD, while the distal 1 kb has orf calC. In silico translation of CalC and subsequent BLAST analysis revealed no homology to known proteins, whereas translation of calD to its protein CalD revealed the presence of three amino acid motifs that are normally expressed in O-methyl transfer The enzyme's S-adenosyl nucleosyl group is conserved in methionein. Therefore, it was assumed that calD was not resp...
Embodiment 3
[0112] Embodiment 3: the expression of protein CalC
[0113] The protein mbp-CalC was overexpressed and purified for further analysis. mbp-CalC was purified to homogeneity from pRE7 / E. coli as identified by SDS-PAGE. Overnight LB culture derived from fresh pRE7 / Escherichia coli colonies (containing 50 mg ml -1 Ampicillin and 50ng ml -1 Calicheamicin) grow at 37°C, 250rpm to A 600 = 0.5, induced with 0.5 mM IPTG, and continued to grow overnight. Cells were harvested (4,000 xg, 4°C, 20 min), resuspended in buffer A (50 mM Tris-Cl, pH 7.2, 200 mM NaCl, 1 mM EDTA), and sonicated. Cell debris was removed by centrifugation (5,000 xg, 4°C, 20 minutes). Apply the supernatant to an amylose affinity column (1.5×7.0cm, 1mL min -1 )superior. The desired mbp-CalC protein was eluted with buffer A containing 10 mM maltose. The eluate was concentrated and chromatographed on an S-300 column (50 mM Tris-Cl, pH 7.5, 200 mM NaCl). The active fraction was used immediately, or stored froze...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com