Method for producing recombinant proteins in micro-organisms
A production method and microbial technology, applied in biochemical equipment and methods, microbial measurement/inspection, peptide/protein components, etc., can solve problems such as high cost of culture conditions, low cost protein yield, and difficult expression of recombinant protein products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1a
[0204] Example 1a: Amplification of the lysine plasminogen gene, the gene is inserted into the codon of the Kex2 restriction site at the 5' end
[0205]Using the QIAGEN plasmid extraction kit (QIAGEN, Hilden), the plasmid pPLGKG was extracted from the HB101 Escherichia coli strain (containing pPLGKG) (Forsgren et al., FEBS Lett. March 23, 1987; 213(2):254-60(2 )), which contains the pre-glutamate-plasminogen (pre-Glu-plasminogen) gene. 150 ng of pPLGKG-DNA was digested with 10 U of EcoRI restriction enzyme (Roche, Mannheim) to linearize it, and then the DNA was purified using QIAquick PCR purification kit (QIAGEN, Hilden). The plasminogen gene was amplified with primers NO34 (Seq. ID No. 1) and NO36 (Seq. ID No. 2). In addition to the base complementary to the plasminogen gene, the oligonucleotide primer NO36 also has the codon of the Kex2 restriction site. Each reaction system contained 0.5U Pwo-DNA-polymerase (Hybaid, Heidelberg), 400 nM oligonucleotide primer, 200 μM dNTP...
Embodiment 1b
[0207] Example 1b: Cloning of the plasminogen gene into the pPICZaA vector
[0208] 400 ng of the PCR product was digested with 10 U each of KspI and XhoI (Roche, Mannheim) restriction enzymes. Plasmid pPICZαAA (Invitrogen, Groningen, Netherlands) contains the pre-propeptide sequence derived from Saccharomyces cerevisiae α-factor, and 300 ng of the DNA of the plasmid was digested with 10 U of KspI and XhoI restriction endonucleases. The digested DNA was separated on 0.9% agarose gel, and the obtained fragments were recovered from the gel with a QIAquick gel extraction kit (QIAGEN, Hilden). The vector DNA and the insert DNA were mixed and ligated overnight at 4° C. with 1 U T4-DNA-ligase (Roche, Mannheim).
[0209] Subsequently, the ligated product DNA was purified using the QIAquick PCR purification kit, and the purified product was used to transform Escherichia coli JM109 by electroporation.
[0210] The JM109 Escherichia coli cells transformed by electroporation were inocu...
Embodiment 1c
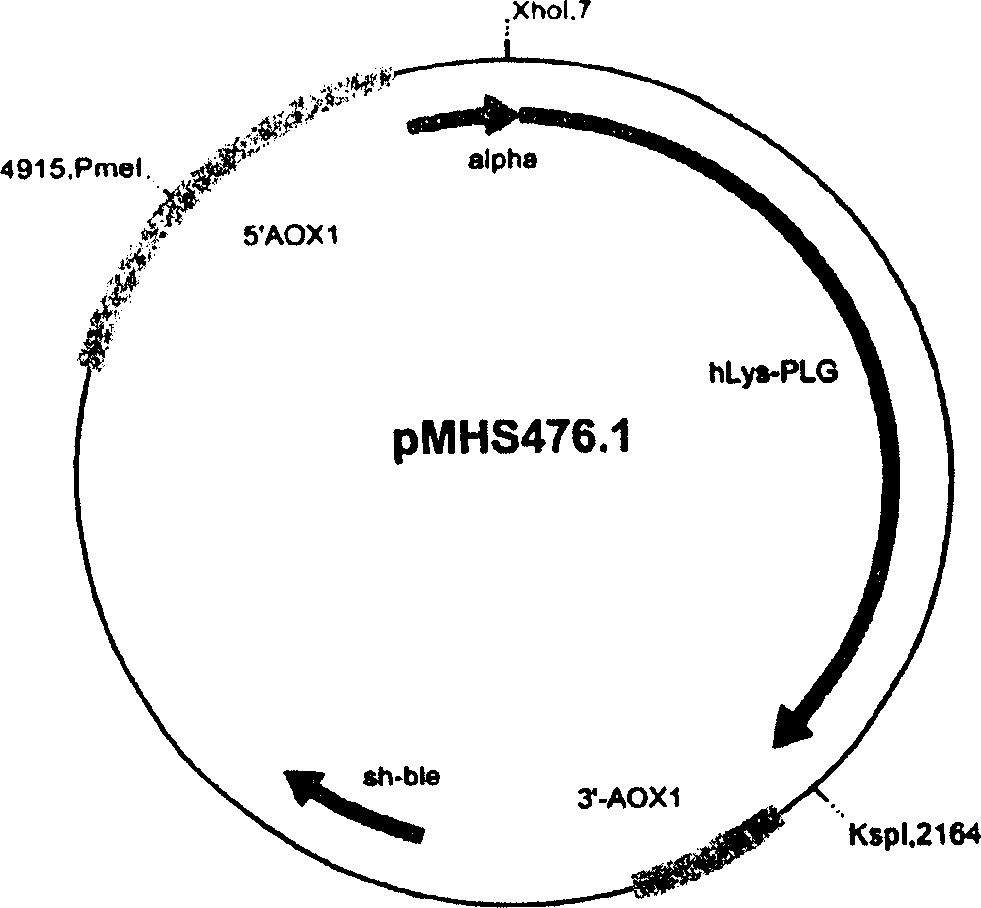
[0212] Example 1c: Transformation of Pichia pastoris with plasmid pMHS476.1
[0213] Using QIAGEN plasmid midi kit (QIAGEN plasmid midi kit, QIAGEN, Hilden) from JM109 Escherichia coli strain (containing pMHS476.1), the expression vector pMHS476.1 plasmid DNA of plasminogen was extracted. Digest 10 μg of pMHS476.1-DNA with 100U PmeI (New England Biolabs, Frankfurt) to linearize it, and follow EasySelect TM According to the method described in the operation manual of the Pichia expression kit, it was electroporated into the Pichia pastoris KM71H strain, and the genotype of the KM71H strain was his4::HIS 4 arg 4 aoxl::Pichia pastoris Y - ARG 4 of strain 11430 (Northern Regional Research Laboratories, Peoria, USA). After the clones were grown in the YPDS solid medium for three to four days, they were plated and cultured in the YPDS solid medium containing 100 μg / ml Meiyilin, and used to inoculate the liquid medium. The clones were named Pichia pastoris KM71H / pMHS476.1-1 / a, whe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com