Soluble fragments of the SARS-cov spike glycoprotein
A technology of fragments and coupled proteins, which can be used in the fields of peptide/protein components, resistance to vector-borne diseases, medical preparations containing active ingredients, etc., and can solve problems such as alveolar wall damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0145] The preparation of monoclonal antibodies is also routine (see Kohler & Milstein, Nature, 256:495 (1975); Coligan et al., sections 2.5.1-2.6.7; and Harlow et al., Antibodies: A Laboratory Manual, page 726 ( Cold Spring Harbor Pub. 1988)), incorporated herein by reference. Simply put, monoclonal antibodies can be injected into mice with a composition containing an antigen, and after the presence of antibodies is confirmed from the collected serum samples, the spleen is taken to obtain B lymphocytes, and the B lymphocytes are fused with myeloma cells to form hybridomas cells, hybridoma cells are cloned, positive clones producing antibodies against the antigen are screened, and antibodies are isolated from the culture medium of the hybridoma cells. Antibodies can be isolated and purified from the culture medium of hybridoma cells by various known methods. These separation techniques include affinity chromatography with Protein A Sepharose, size exclusion chromatography, an...
Embodiment 1
[0282] Cloning of the spike protein
[0283] The nucleic acid sequence encoding the full-length Spike protein was obtained by overlapping polymerase chain reaction (PCR). Overlapping clones containing the spike protein were obtained from the British Columbia Cancer Agency (Vancouver, British Columbia). The following primers were used in a PCR reaction to amplify the nucleic acid sequence encoding the full-length SARS-CoV Spike protein: Clone 1: Forward primer: 5′-AGTC GGA TCC GGT AGG CTT ATC ATT AGA G-3' (SEQ ID NO: 3); reverse primer: 5'-CCA TCA GGG GAG AAA GGC AC-3 (SEQ ID NO: 4). Clone 2: forward primer: 5'-GTG CCT TTC TCC CCT GAT GG-3' (SEQ ID NO: 5); reverse primer: 5'-GAA GAG CAG CGC CAG CAC C-3' (SEQ ID NO: 6). Clone 3: Forward primer: 5'-GGT GCT GGC GCT GCT CTT C-3' (SEQ ID NO: 7); Reverse primer: 5'-A CTG TCT AGA GTT CGT TTA TGT GTAATG-3 (SEQ ID NO: 8).
[0284] The nucleic acid fragment generated by overlapping PCR amplification of the nucleic acid fragment b...
Embodiment 2
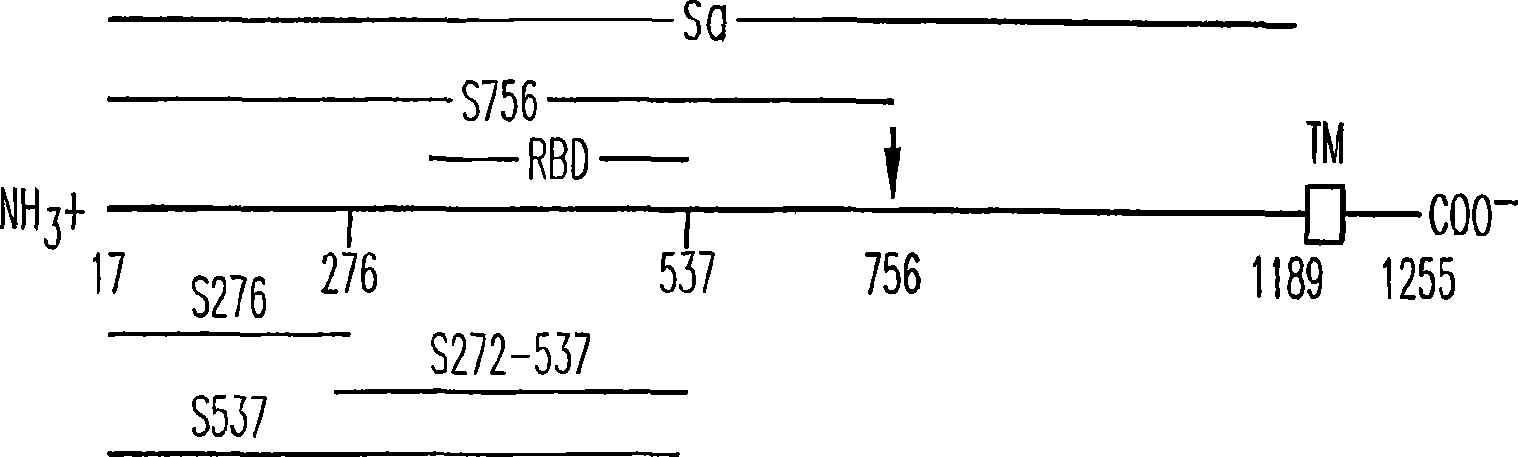
[0287] Generation of amino-terminal (S1) and carboxy-terminal (S2) fragments of the full-length Spike protein
[0288] Computer analysis was used to determine the potential functional segmentation points of the amino-terminal (S1) and carboxy-terminal (S2) of the spike protein, and the segmentation points between S1 and S2 were at 758 and 761 of SEQ ID NO: 1 ( 758 RNTR 761 )between. The method of PCR was used to prepare the amino-terminal fragment (S1) and the carboxy-terminal (S2) fragment encoding the spike protein.
[0289] The following primer pair, S1 forward primer: 5′-AGTC GGA TCC GAC CGG TGCACC ACT TTT G-3' (SEQ ID NO: 9), and S1 reverse primer: 5'-AGTC GGG CCC CTG TTC AGC AGC AAT ACC-3' (SEQ ID NO: 10) was used to prepare a nucleic acid fragment encoding amino acid residues 17 to 757 of the Spike protein. Two restriction sites, BamHI and ApaI (underlined parts of the two primer sequences), were used to clone the nucleic acid fragment (S1) encoding the N-termi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com