Method of preparation of pharmaceutically grade plasmid DNA
A plasmid-level technology, applied in the production and separation of pharmaceutical-grade plasmid DNA, can solve the problems of difficulty in plasmid DNA and inability to obtain nucleic acids in large quantities
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0208] The adjustment of diameter to flow rate used in the coils of the continuous lysis system is based on the calculation of Reynolds numbers. Because the following analysis assumes Newtonian behavior of the fluid, the numbers reported below are only fully valid in B1a and to some extent in B2.
[0209] The value of the Reynolds number allows one skilled in the art to determine the type of behavior encountered. Here, we will only focus on fluid flow in pipes (hydraulic engineering).
[0210] 1) Non-Newtonian fluid
[0211] The two most commonly encountered non-Newtonian fluids in industry are Bingham and Ostwald de Waele.
[0212] In this case, the Reynolds number (Re) is calculated as follows:
[0213] ReN is the generalized Reynolds number
[0214] ReN=(1 / (2 n-3 ))×(n / 3n+1) n ×((ρ×D n ×w 2-n ) / m) (1)
[0215] D: Inner diameter of the cross section (m)
[0216] ρ: volume mass of fluid (volum nass) (kg / m 3 )
[0217] w: space velocity of the fluid (m / s)
[0218]...
Embodiment 2
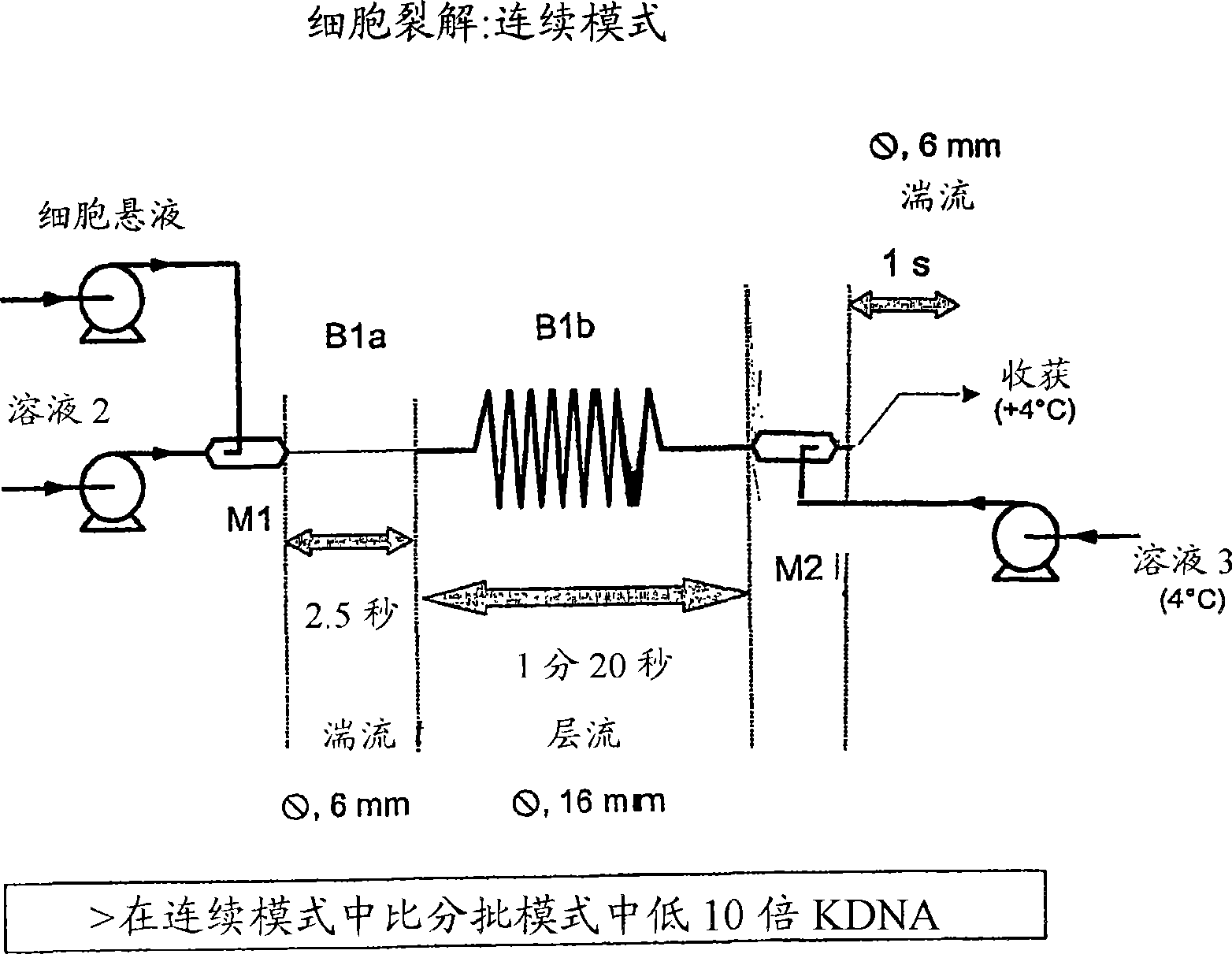
[0258] We can decompose the CL system into 5 steps: In a specific implementation, the structure is as follows:
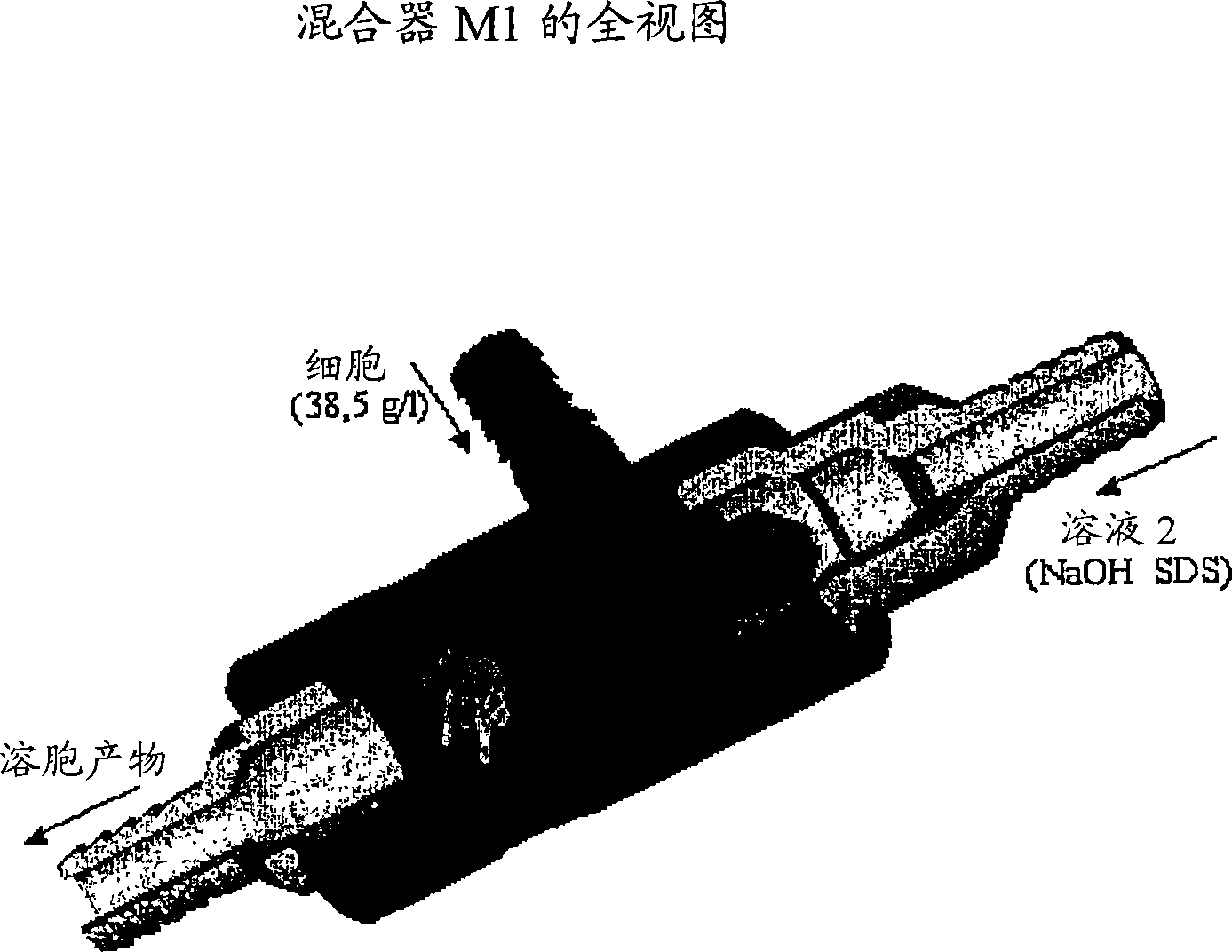
[0259] 1) Mixing: cells (in solution 1) + solution 2 (M1 + 3m of 6mm tubing). Begin cell lysis by SDS, there is no risk of fragmentation as long as the DNA is not denatured.
[0260] 2) End of lysis and denaturation of gDNA (13m, 16mm tubing).
[0261] 3) Mixing: lysate + solution 3 (M2 + 3m, 6mm tubing).
[0262] 4) Harvest the neutralized lysate at 4°C
[0263] 5) Sediment flocs and large gDNA fragments overnight at 4°C
[0264] The following conditions can be used to perform sequential lysis:
[0265] - Solution 1: EDTA 10 mM, Glucose (Glc) 9 g / l and Tris HCl 25 mM, pH 7.2.
[0266] - Solution 2: SDS 1% and NaOH 0.2N.
[0267] - Solution 3: acetic acid 2M and potassium acetate 3M.
[0268] - Flow rate 60 l / h: solution 1 and solution 2
[0269] - Flow rate 90 l / h: solution 3.
[0270] - The cells were adjusted to 38.5 g / l with solution 1.
[0271] Cells ...
Embodiment 3
[0287] The column used was a 1 ml HiTrap column, activated with NHS (N-hydroxysuccinimide, Pharmacia), connected to a screw-type compression pump (output < 1 ml / min. The specific oligonucleotide used had NH2 at the 5' end The group, the sequence of which is as follows: 5'-GAGGCTTCTTCTTCTTCTTCTTCTT-3' (SEQ ID NO: 1)
[0288] The buffers used in this example are as follows:
[0289] Coupling buffer: 0.2M NaHCO3, 0.5M NaCl, pH 8.3.
[0290] Buffer A: 0.5M ethanolamine, 0.5M NaCl, pH 8.3.
[0291] Buffer B: 0.1M acetic acid, 0.5M NaCl, pH 4.
[0292] The column was washed with 6 ml of 1 mM HCl, then the oligonucleotide diluted in coupling buffer (50 nmol in 1 ml) was applied to the column for 30 minutes at room temperature. The column was washed three times with 6 ml buffer A followed by 6 ml buffer B. The oligonucleotides are thus covalently bound to the column via CONH linkages. The column is stored at 4°C in PBS, 0.1% NaN3 and can be used at least 4 times.
[0293] The fo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com