Human myocardial troponin I subunit nucleic acid adaptor and its application
A cardiac troponin, subunit nucleic acid technology, applied in the biological field, can solve the problems of interference factors, difficult to establish cTnI standardization, different antibody titers, etc., and achieve high affinity and specificity, repeatability and stability. The effect of good and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] A human cardiac troponin I subunit nucleic acid aptamer, characterized in that it has the nucleotide sequence described in SEQ ID No.1 in the sequence listing or has the nucleotide sequence described in SEQ ID No.2 in the sequence listing Sequence or the nucleotide sequence described in SEQ ID No.3 in the Sequence Listing.
[0031] The nucleotide sequence described in SEQ ID No.1:
[0032] cccctgcagg tgattttgct caagtcgaga gcggtgcatc tagggtctct agctcgggaa
[0033] agtatcgcta atcaggcgga t 81
[0034] Nucleotide sequence described in SEQ ID No.2:
[0035] cccctgcagg tgattttgct caagtgctta atcgagggta tcgtggggca gttgggaggg 60
[0036] agtatcgcta atcaggcgga t 81
[0037]Nucleotide sequence described in SEQ ID No.3:
[0038] cccctgcagg tgattttgct caagtgccgt caacatgtcc tagtaggggt ctcaggggtg 60
[0039] agtatcgcta atcaggcgga t 81
[0040] The single-stranded DNA random library and primers used in SELEX in the present invention are synthesized by Invitrogen, with fixed sequ...
Embodiment 2
[0042] Screening and preparation of a human cardiac troponin I subunit nucleic acid aptamer
[0043] Preparation of ssDNA library by PCR
[0044] The PCR reaction system is:
[0045] 10× amplification buffer 10ul
[0046] 4 kinds of dNTP mixture each 200umol / L
[0047] Primer 1 100pmol (5'CCCCT GCAGGTGATTTTGTCCAAGT 3')
[0048] Primer 2 100pmol (5'-biotin-ATCCGCCTGA TTAGCGAT ACT3')
[0049] Template DNA lug (5’CCCCTGCAGGTGATTTT GCTCAA GT-(N35)-AGTATCGCTAATCAGGCGGAT 3’)
[0050] Taq DNA Polymerase 2.5u
[0051] Mg 2+ 1.5mmol / L
[0052] Add double or triple distilled water to 100ul
[0053] The PCR reaction conditions were: pre-denaturation at 94°C for 3 min, then denaturation at 94°C for 40 s, annealing at 65°C for 1 min, extension at 72°C for 2 min, and finally extension at 72°C for 7 min.
[0054] A dsDNA library with biotin at the 3' end was amplified by PCR, and the dsDNA product was separated and purified, and combined with streptavidin magnetic beads; ...
Embodiment 3
[0057] Gel retardation assay to detect the binding of a human cardiac troponin I subunit nucleic acid aptamer to cTn I
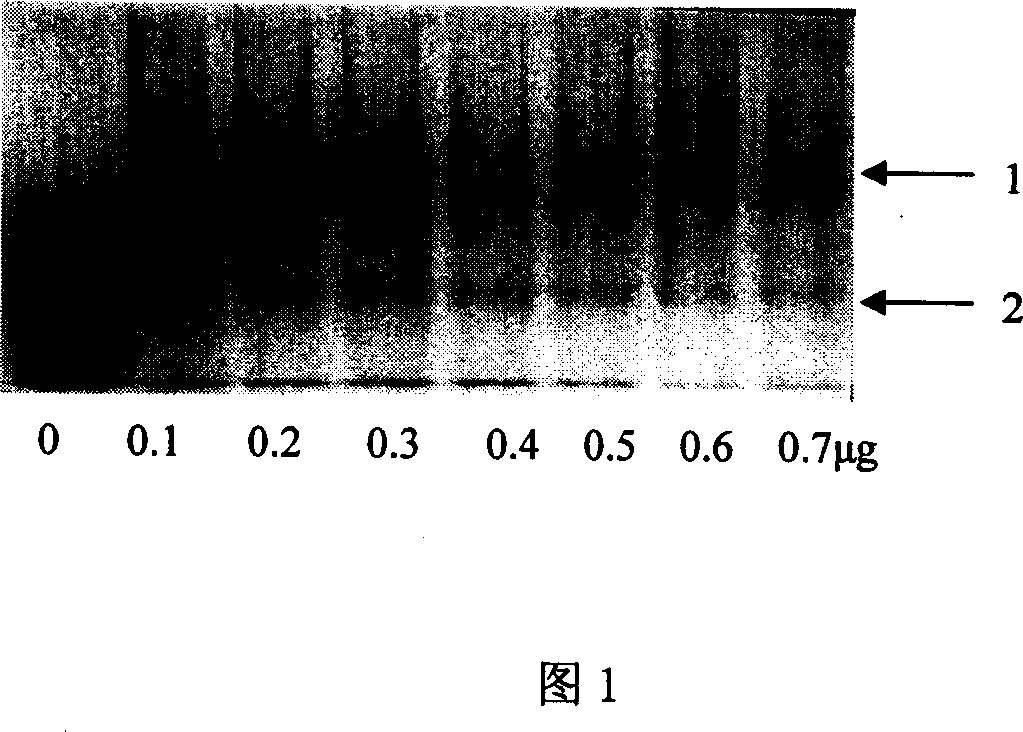
[0058] The obtained human cardiac troponin I subunit nucleic acid aptamer (nucleotide sequence described in SEQ ID No.1 in the sequence listing) was denatured at 95° C. for 5 minutes, quickly ice-water bathed for 5 minutes, and 10 μl of binding buffer was added. Add different amounts of cTnI (0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7 μg), and keep at 37° C. for 30 minutes (water bath). Load the sample to 5% polyacrylamide gel, electrophoresis, and observe the results by silver staining. As shown in Figure 1, gel hysteresis occurs after a human cardiac troponin I subunit nucleic acid aptamer binds to cTnI. No gel hysteresis without protein.
[0059] Different amounts of 0 μg, 0.1 μg, 0.2 μg, 0.3 μg, 0.4 μg, 0.5 μg, 0.6 μg, 0.7 μg of cTn I were incubated with 100 pmol of a human cardiac troponin I subunit nucleic acid aptamer for non-denaturation Polyacrylamide ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com