Method for typing of HLA class I alleles
a typing method and allele technology, applied in the field of methods, reagents and kits for typing of hla class i alleles, can solve the problems of reducing the credibility of testing results, affecting the quality of testing, and the collection of specific antibodies, and requiring complicated manipulation of methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
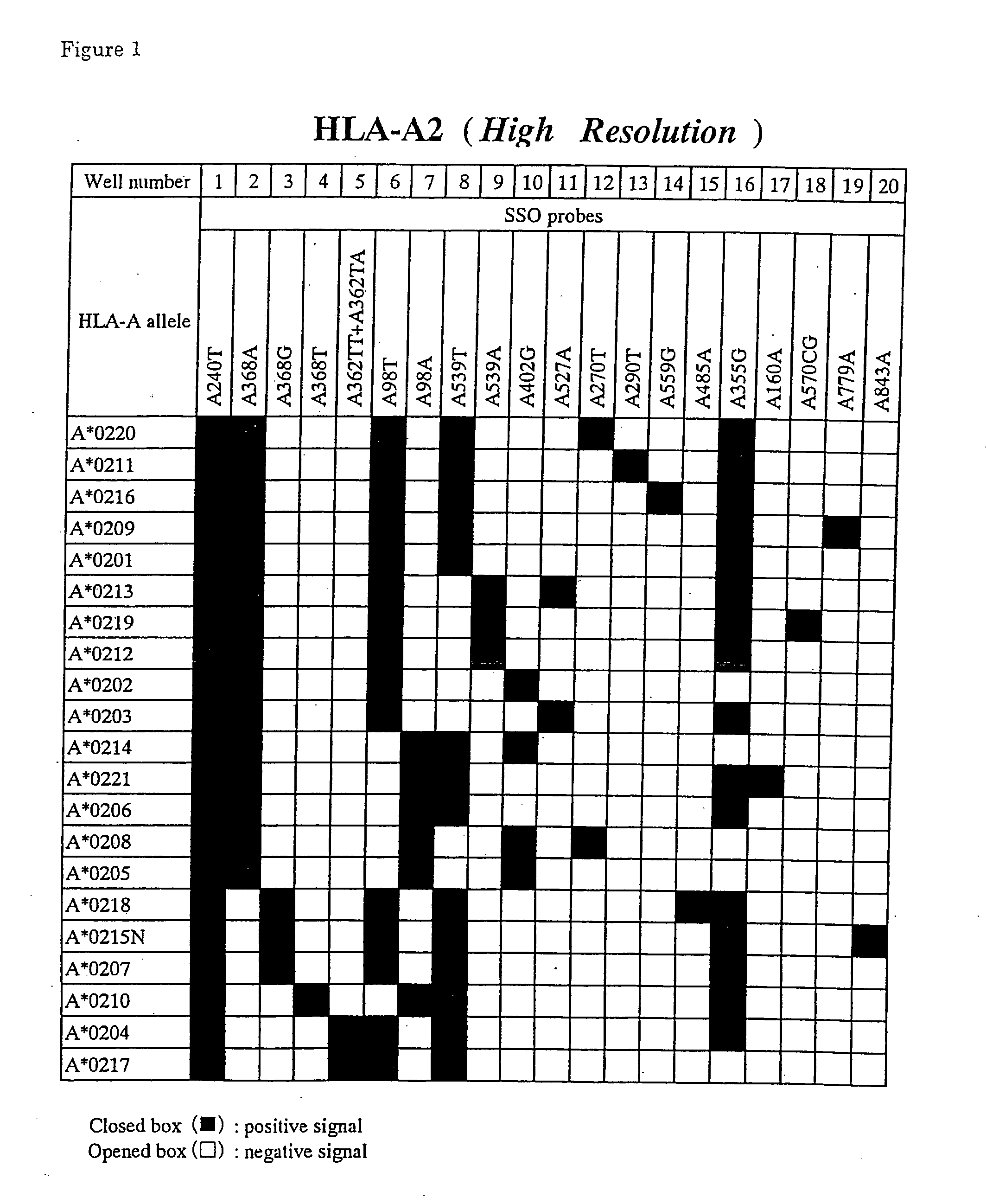
[0064] Leukocytes (Samples 1-4) which were isolated from peripheral blood (about 10 ml) of normal subjects according to usual methods, were lysed in 500 μl of guanidine thiocyanate buffer (4M guanidine thiocyanate, 25 mM sodium citrate(pH7.0), 0.5% sodium N-lauroylsarcosinate, 1% mercaptoethanol). The solution was extracted twice with phenol to eliminate proteins. After mixing with 3M sodium acetate buffer (pH 5.2), genome DNAs were obtained by adding twice volume of chilled ethanol. By using this DNAs, typing of the HLA-A2 alleles was performed as follows.
[0065] By using A2-5T and 5′-biotinylated A3-273T for a primer pair, amplification of the region containing the exon 2, the intron 2 and the exon 3 of the HLA-A2 alleles from DNAs described above was performed by the PCR method. Likewise, by using A4-8C and 5′-biotinylated A4-254G for a primer pair, amplification of the region containing the exon 4 of the HLA-A alleles was also performed by the PCR method. Th...
example 2
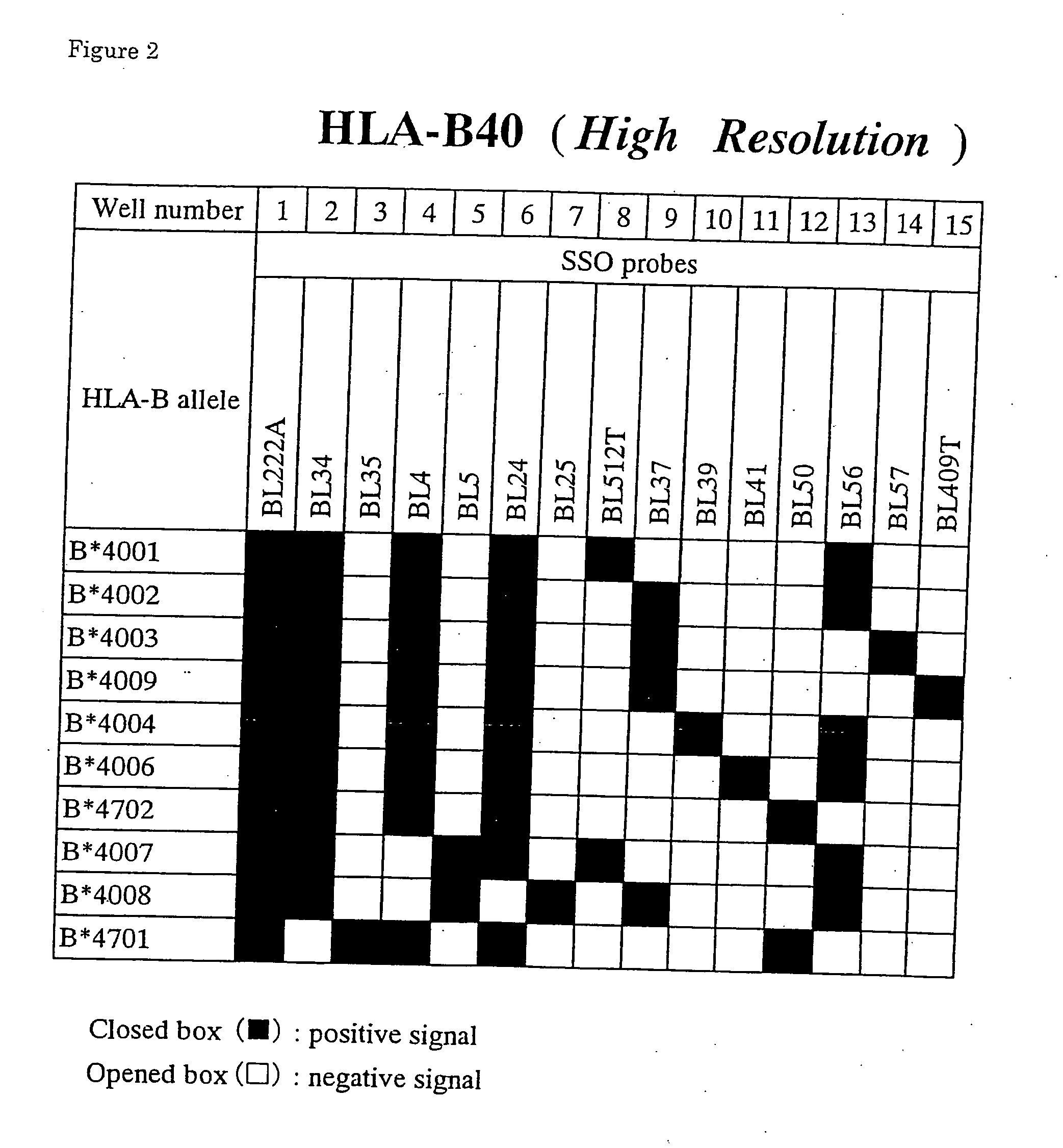
[0069] Leukocytes (Samples 5-8) which were isolated from peripheral blood (about 10 ml) of normal subjects according to usual methods, were lysed in 500 μl of guanidine thiocyanate buffer(4M guanidine thiocyanate, 25 mM sodium citrate(pH7.0), 0.5% sodium N-lauroylsarcosinate, 1% mercaptoethanol). The solution was extracted twice with phenol to eliminate proteins. After mixing with 3M sodium acetate buffer (pH 5.2, genome DNAs were obtained by adding twice volume of chilled ethanol. By using this DNAs, typing of the HLA-B40 alleles was performed as follows.
[0070] By using BASF-1 and 5′-biotinylated BASR-1 for a primer pair, amplification of the region containing the exon 2, the intron 2 and the exon 3 of the HLA-B40 alleles from DNAs described above was performed by the PCR method. The reaction solution was composed of genomic DNAs (100 ng), 1.4 units of thermostable DNA which was pretreated with Taq Start™Antibody for 5 min at room temperature, 33.5 mM Tris-HC...
example 3
HLA-A Antigen and Allele Typing
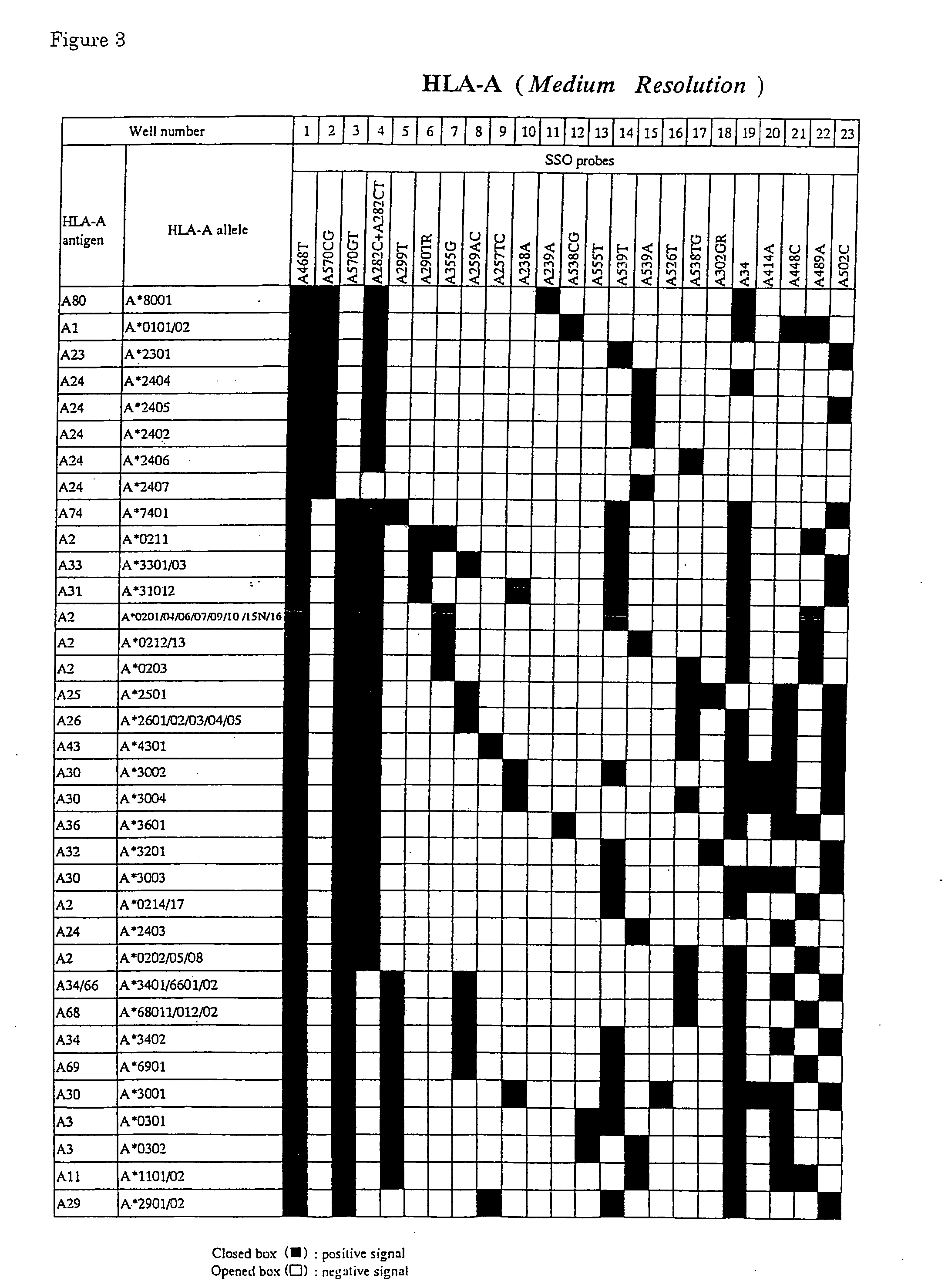
[0074] Leukocytes (Samples 9-12) which were isolated from peripheral blood (about 10 ml) of normal subjects according to usual methods, were lysed in 500 μl of guanidine thiocyanate buffer (4M guanidine thiocyanate, 25 mM sodium citrate(pH7.0), 0.5% sodium N-lauroylsarcosinate, 1% mercaptoethanol). The solution was extracted twice with phenol to eliminate proteins. After mixing with 3 M sodium acetate buffer (pH5.2), genome DNAs were obtained by adding twice volume of chilled ethanol. By using this DNAs, typing of the HLA-A antigens and alleles was performed as follows.
[0075] By using CGA011, CGA012 and 5′-biotinylated AIn3-66C for a primer pair, amplification of the region containing the exon 2, the intron 2 and the exon 3 of the HLA-A alleles from DNAs described above was performed by the PCR method. The reaction solution was composed of genomic DNAs (100 ng), 1.4 units of thermostable DNA polymerase which was pretreated with Taq Start™Antibody for...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com