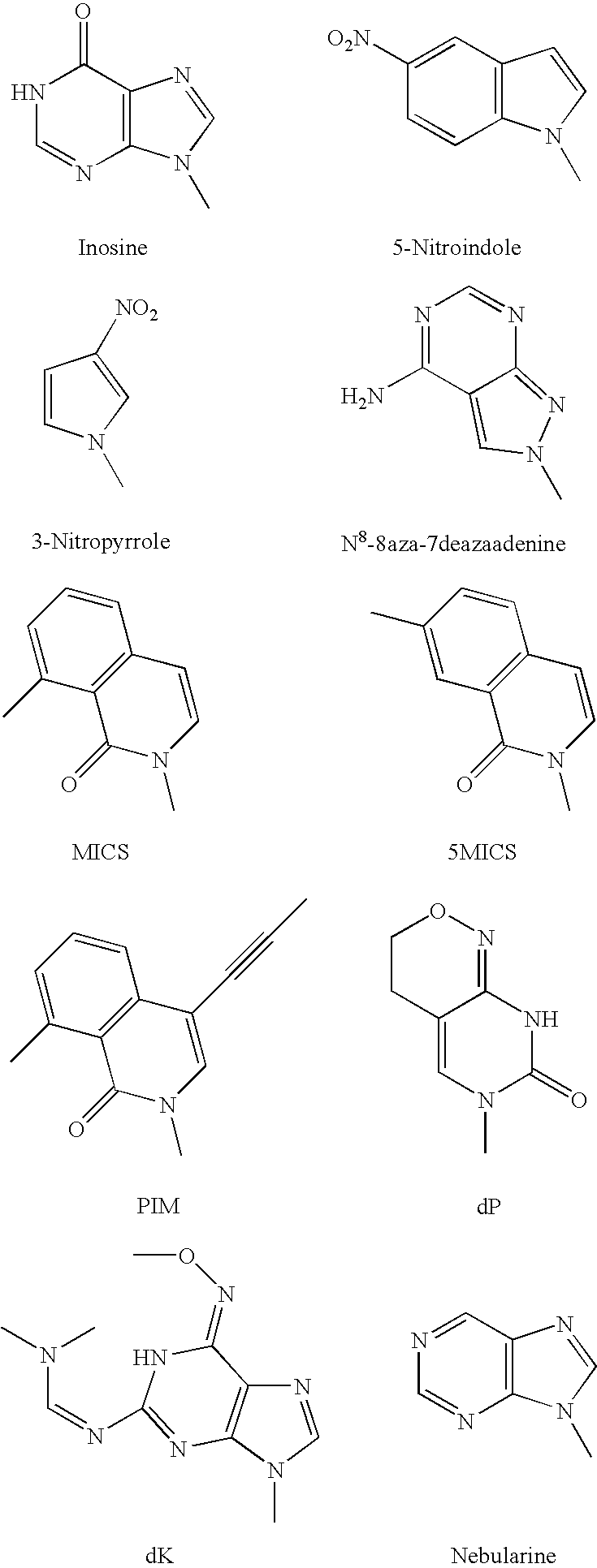
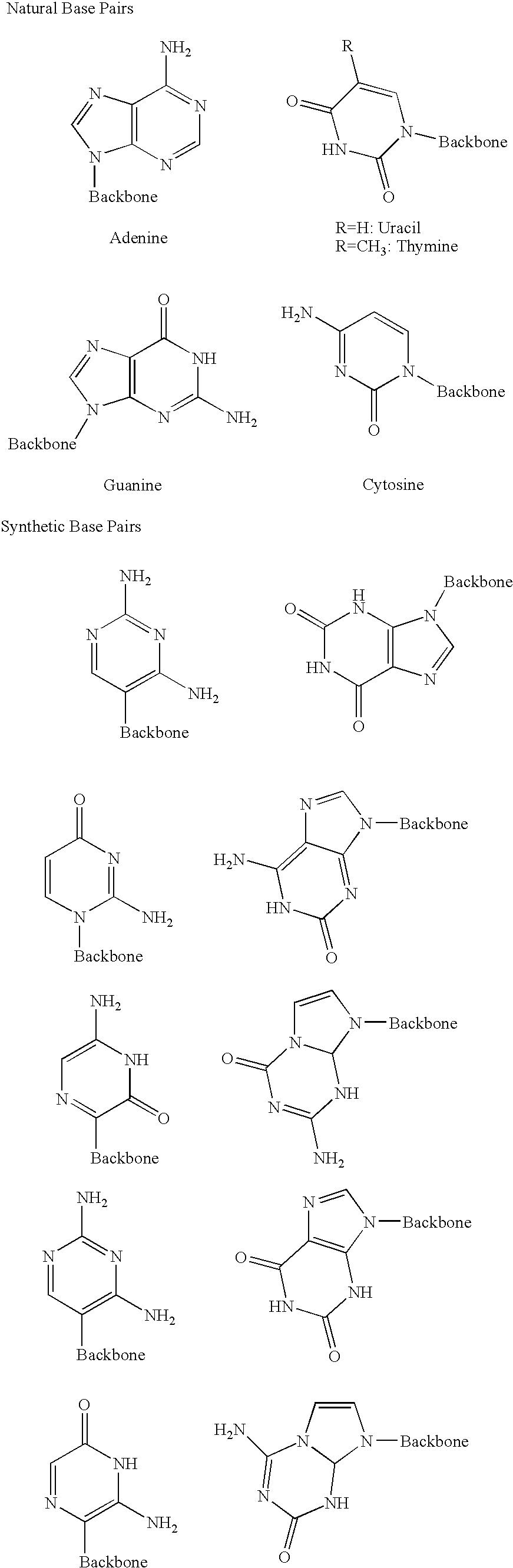
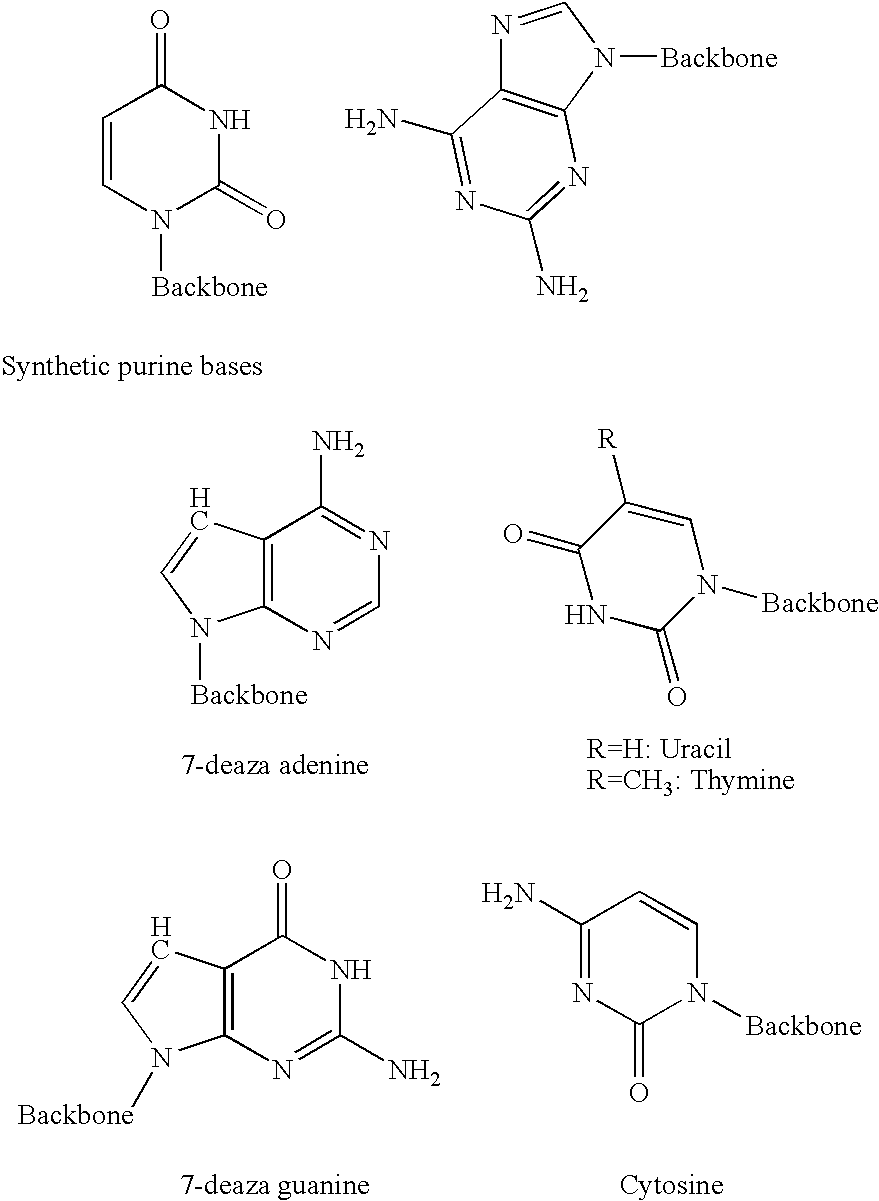
Method for synthesising templated molecules
a templated molecule and synthesis method technology, applied in the direction of nucleotide libraries, electric generator control, machines/engines, etc., can solve the problems of cumbersome split-and-combine methods, difficult generation of molecules carrying new properties, and inability to achieve one-pot amplification of library members, etc., to achieve a higher ratio of complexes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
[0271] Mix 2 μl Buffer A, 2 μl relevant oligo 1 (0.2 pmol / ul), 1 μl relevant oligo 2 (10 pmol / ul), 1 μl relevant oligo 3 (10 pmol / ul), 4 μl H2O. (See table II, below)
TABLE IIOligo 1 (32P-Experimentlabelled)Oligo 2Oligo 3GAh 5Ah 6NoneHAh 5Ah 6Ah 7IAh 5Ah 6Ah 8JAh 5Ah 6Ah 9KAh 5Ah 6Ah 11
[0272] Anneal as described above. Add 1 μl 100 mM, 10 mM or 1 mM TSAT (Tris-succinimidyl aminotriacetate, Pierce cat#33063 dissolved in DMSO). Incubate at 25° C. for about 5 h, then run 10% urea polyacrylamide gel, as described above.
[0273] The results are shown in FIG. 2
example 3
[0274] Mix 2 μl Buffer A, 2 μl relevant oligo 1 ( 0.2 pmol / ul), 1 μl relevant oligo 2 (10 pmol / ul), 1 μl relevant oligo 3 (10 pmol / ul), 4 μl H2O. (See table III, below)
TABLE IIIOligo 1 (32P-Experimentlabelled)Oligo 2Oligo 3LAh 1Ah 6NoneMAh 1Ah 6Ah 7NAh 1Ah 6Ah 8OAh 1Ah 6Ah 9PAh 1Ah 6Ah 11
[0275] Anneal as described above. Add 1 μl 1M, 100 mM, 10 mM or 1 mM EDC (1-Ethyl-3-(3-dimethylaminopropyl) Carbodiimide Hydrochloride, Fluka #03450) and 1 μl 100 mM NHS (N-Hydroxysuccinimid) (Aldrich cat #13.067-2). Incubation at 25° C. for about 5 h, and analyze by 10% urea polyacrylamide gel electrophoresis, as described above.
[0276] The results are shown in FIG. 3.
example 4
[0277] Mix 2 μl buffer A, B, C, D, E or F, 2 μl relevant oligo 1 ( 0.2 pmol / ul), 1 μl relevant oligo 2 (10 pmol / ul), 1 μl relevant oligo 3 (10 pmol / ul), 4 μl H2O. ( See table IV, below)
TABLE IVOligo 1 (32P-Experimentlabelled)Oligo 2Oligo 3QAh 1Ah 6Ah 7RAh 5Ah 6Ah 7
[0278] Anneal as described above. Experiment Q is added 1 μl 100 mM EDC and 1 μl 100 mM NHS . Experiment R is added 1 μl 100 mM TSAT. Incubate at 25° C. for about 1.5 h, and then analyze by 10% urea polyacrylamide gel electrophoresis.
[0279] The results are shown in FIG. 4.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com