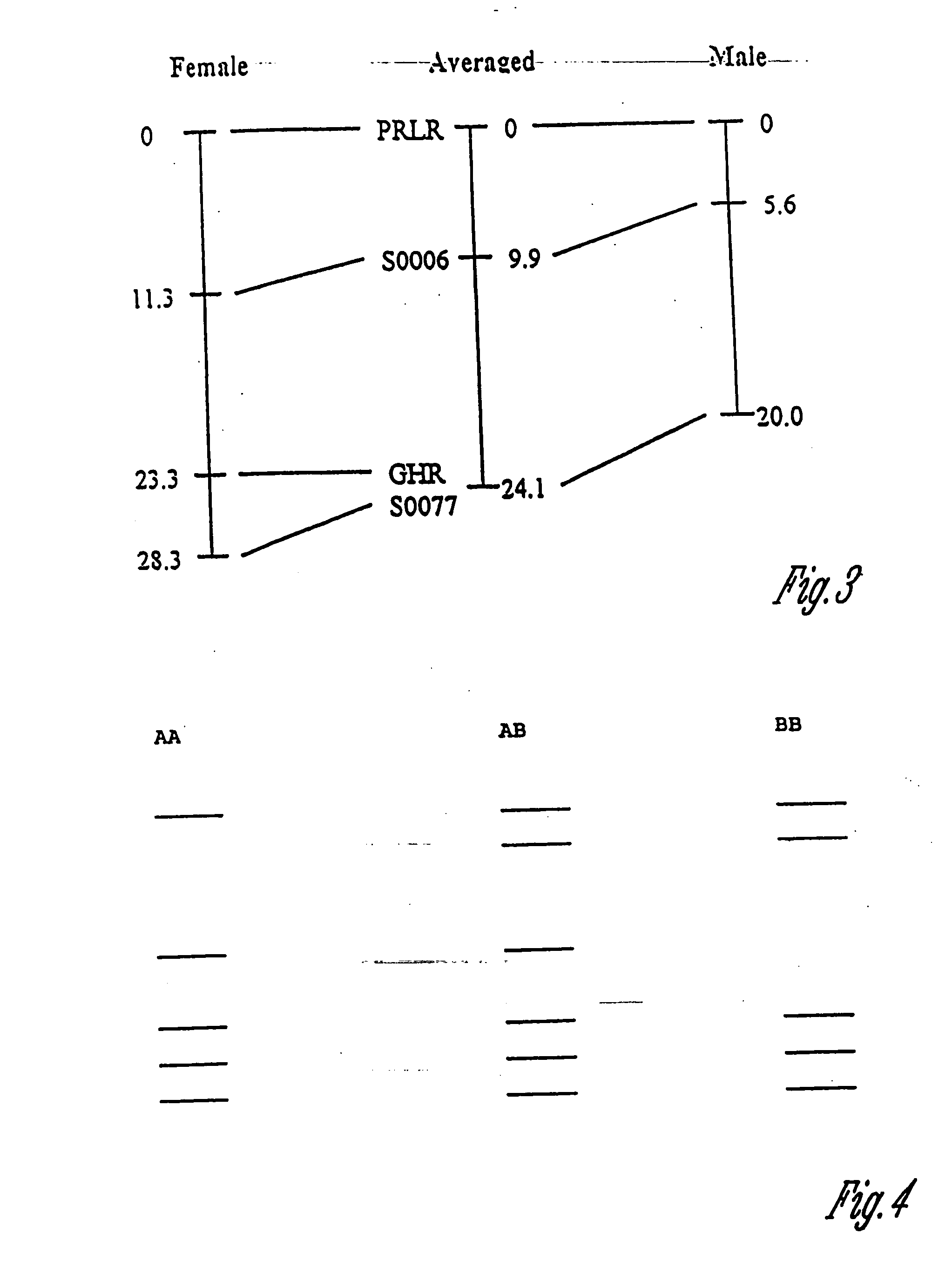
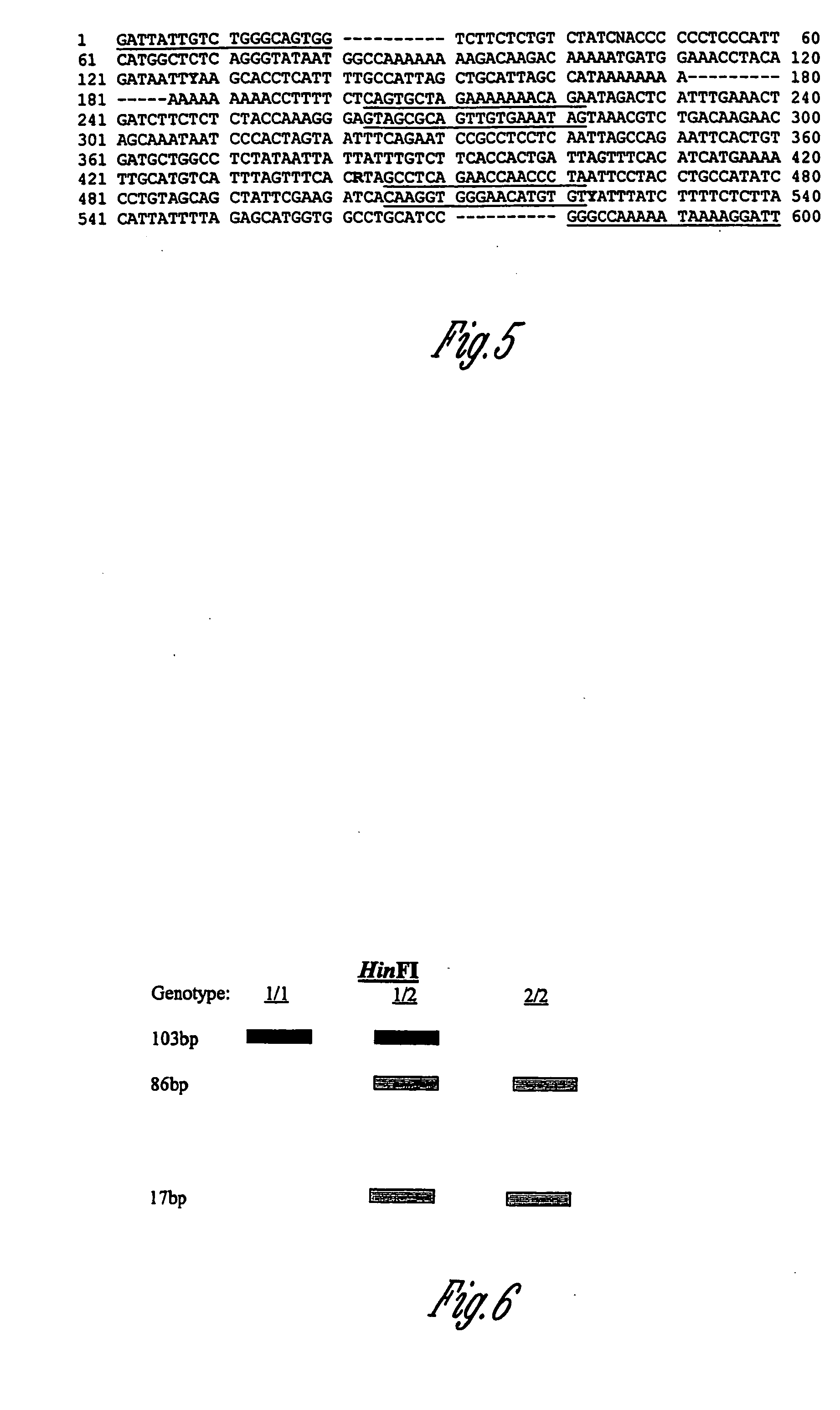
Prolactin receptor gene as a genetic marker for increased litter size in animals
a technology of prolactin receptor and gene, which is applied in the field of genetic differences for reproductive efficiency among pigs, can solve the problems of low heritability of litter size, major limiting factor in the efficient production of pork, and reproductive efficiency,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0083] Due to their high sequence homology and similarity in transcript processing, human (Boutin et al. 1989) and rabbit (Edery et al. 1989) cDNA sequences encoding the prolactin receptor were used to design degenerate primers overlapping the 31 coding and untranslated region. The primers amplified a fragment of approximately 500 base pairs in pig genomic DNA samples and human control. The forward primer 5′-TCA CAA GGT CAA C / TAA AGA TG-3′ (SEQ ID NO:4) and the reverse primer 5′-TGG / A AGA AAG / A AGG CAA G / ATG GT-3′ (SEQ ID NO:5)were used in the following PCR conditions: 93° C. for 3 minutes, 6 cycles of 93° C. 30 seconds, 47° C. 2 minutes, 72° C. 3 minutes, 36 cycles of 93° C. 30 seconds, 53° C. 2 minutes, 72° C. 5 minutes, and a final 72° C. 5 minutes. The Taq polymerase was added last while samples were held at 80° C.
[0084] Fragments from two animals were purified and sequenced in forward and reverse directions. The pig sequence from the coding region was translated to amino acids...
example 2
[0085] PCR TEST for Prolactin Receptor Genetic Marker The PCR amplification test was optimized with the following parameters.
[0086] Primers:
(SEQ ID NO:1)forward primer 5′-CCCAAAACAGCAGGAGAACG-3′(SEQ ID NO:2)reverse primer 5′-GGCAAGTGGTTGAAAATGGA-3′
[0087] PCR Conditions:
Cocktail Mix25uL reaction10X PCR buffer (Promega)2.5uL25 mM MgCl2 (Promega)2.0uL10 mM dNTP's (Boehringer Mannheim)0.5uL20 pmol / uL forward primer0.5uL20 pmol / uL reverse primer0.5uLdd Sterile H2O17.5uL12.5 ng / uL DNA1.5uLTaq Polymerase (Promega)0.125uL
[0088] The first six reagents should be mixed and 18.5 uL of this pre-mix added to each reaction tube. Add the DNA next and then overlay with a drop of sterile mineral oil. Place the reaction tubes on the terminal cycler held at 80° C. Mix the Taq with the remaining cocktail and add 5 uL to the reaction tubes, making sure to submerge the tip beneath the oil.
Thermal Cycler Program:
[0089] 1. 93° C. 3 minutes [0090] 2. 93° C. 30 seconds [0091] 3. 60° C. 1 minute [0092]...
example 3
Association of Genotype with Litter Size
[0101] The PCR test was run as detailed in Example 2 on several sows from Pig Improvement Company, (PIC) . The animals used were PIC line 19 sows which farrowed within a six month period and gilts which were born during this period that would be kept as breeding stock. Blood or tissue samples were collected and shipped to the laboratory where the DNA was extracted and used in the PRLR PCR test. Females had one to three records used in the analysis. Estimated Breeding Value for Total Number Born (BV TNB) was estimated using a mixed linear model where each successive parity of the sow is treated as a repeated record. Only the first three parities of a sow were used. The model includes the covariate of age at farrowing nested within parity, fixed effects of parity, service type (natural or AI), farm-month farrowed, and random permanent environmental and animal effects. Current h2 is assumed as 0.10 and repeatability as 0.21. Average Number Born ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com