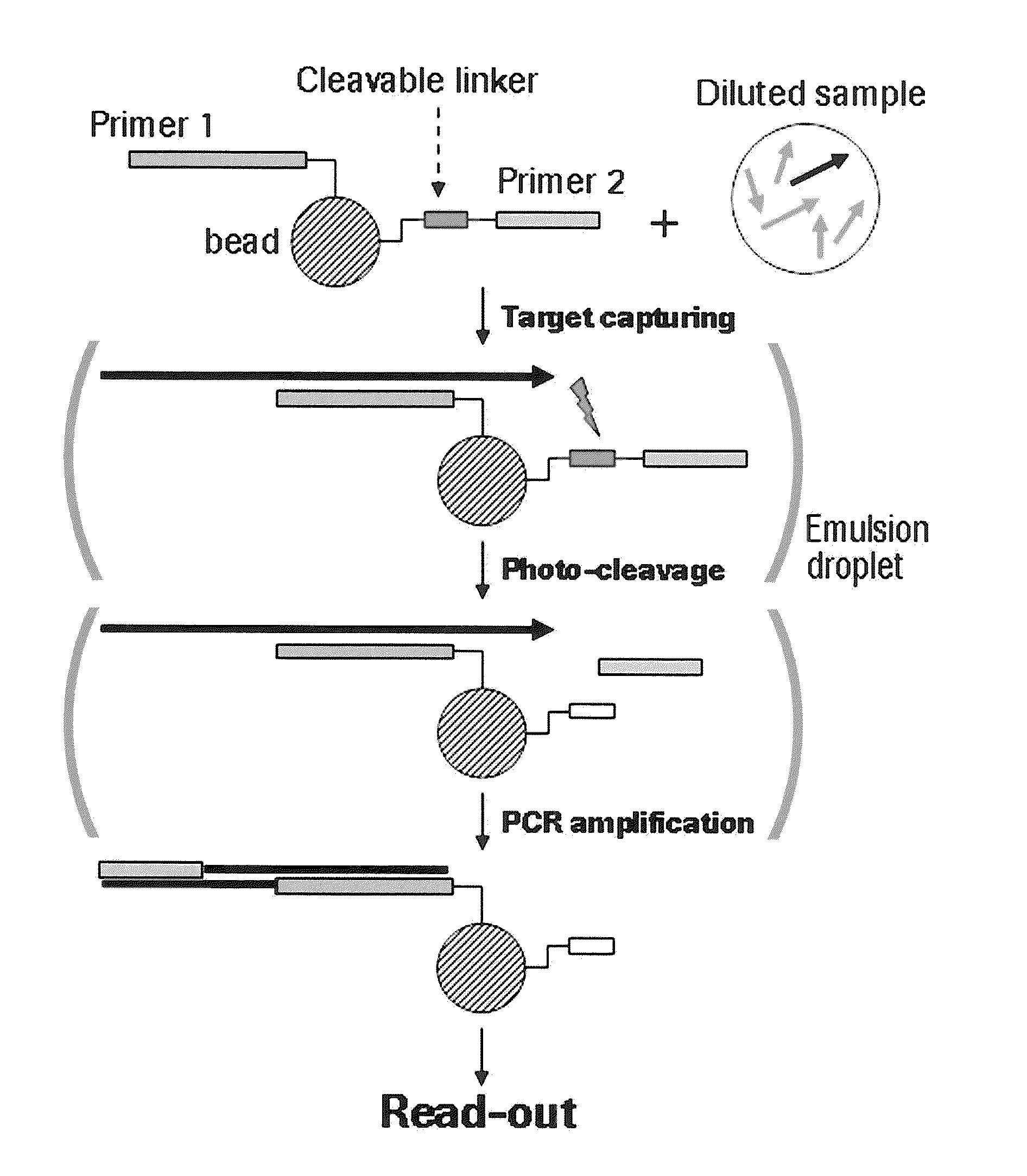
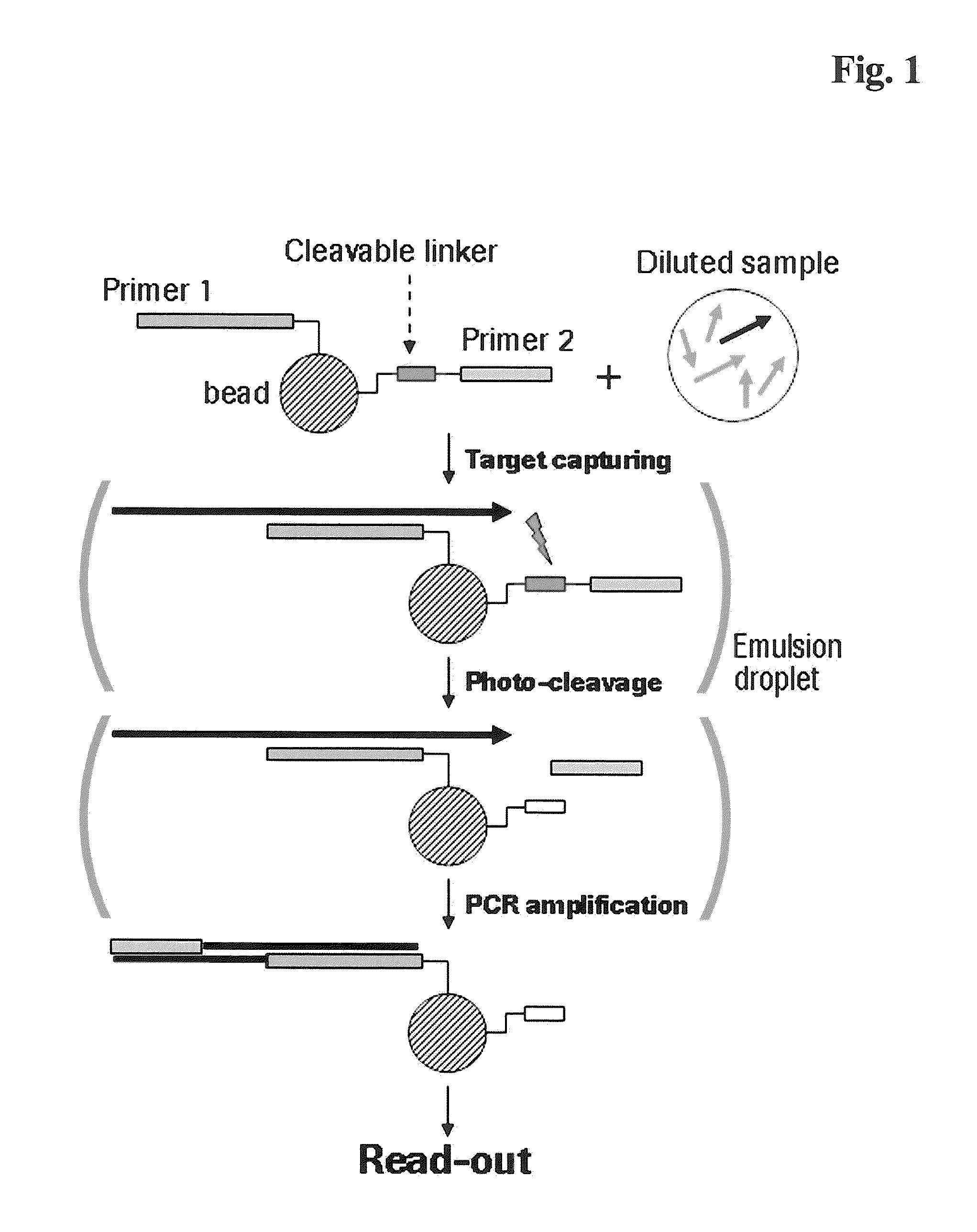
Miniaturized, high-throughput nucleic acid analysis
a nucleic acid analysis and high-throughput technology, applied in the field of nucleic acid analysis, can solve the problem of difficult sample processing and achieve the effect of difficult sample processing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of Primers and Photocleavable Primers
[0194]Oligonucleotide synthesis was carried out on a 4 times 1 μmol scale on an ABI 394 synthesizer. Commercially available tac CPG (Proligo) was used as the support material. All other chemicals for the standard synthesis were obtained from Glen Research. Phosphoramidites with tert. butylphenoxy-acetyl protective groups (known as “tac” or “Expedite” monomers) from Proligo were used. As capping reagent tertbutylphenoxyacetyl acetic anhydride (tac2O) in tetrahydrofuran was used.
[0195]The following commercially available modifiers were used:[0196]5′ amino modifier C6: (6-(4-Monomethoxytritylamino)hexyl-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite[0197]Spacer phosphoramidite 18 (18-O-Dimethoxytritylhexaethyleneglycol, 1-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite[0198]Photocleavble spacer [4-(4,4′-Dimethoxytrityloxy)butyramidomethyl)-1-(2-nitrophenyl)-ethyl]-2-cyanoethyl-(N,N-diisopropyl)-phosphoramidite[0199]BiotindT phosphor...
example 2
Preparation of Beads and Photocleavage
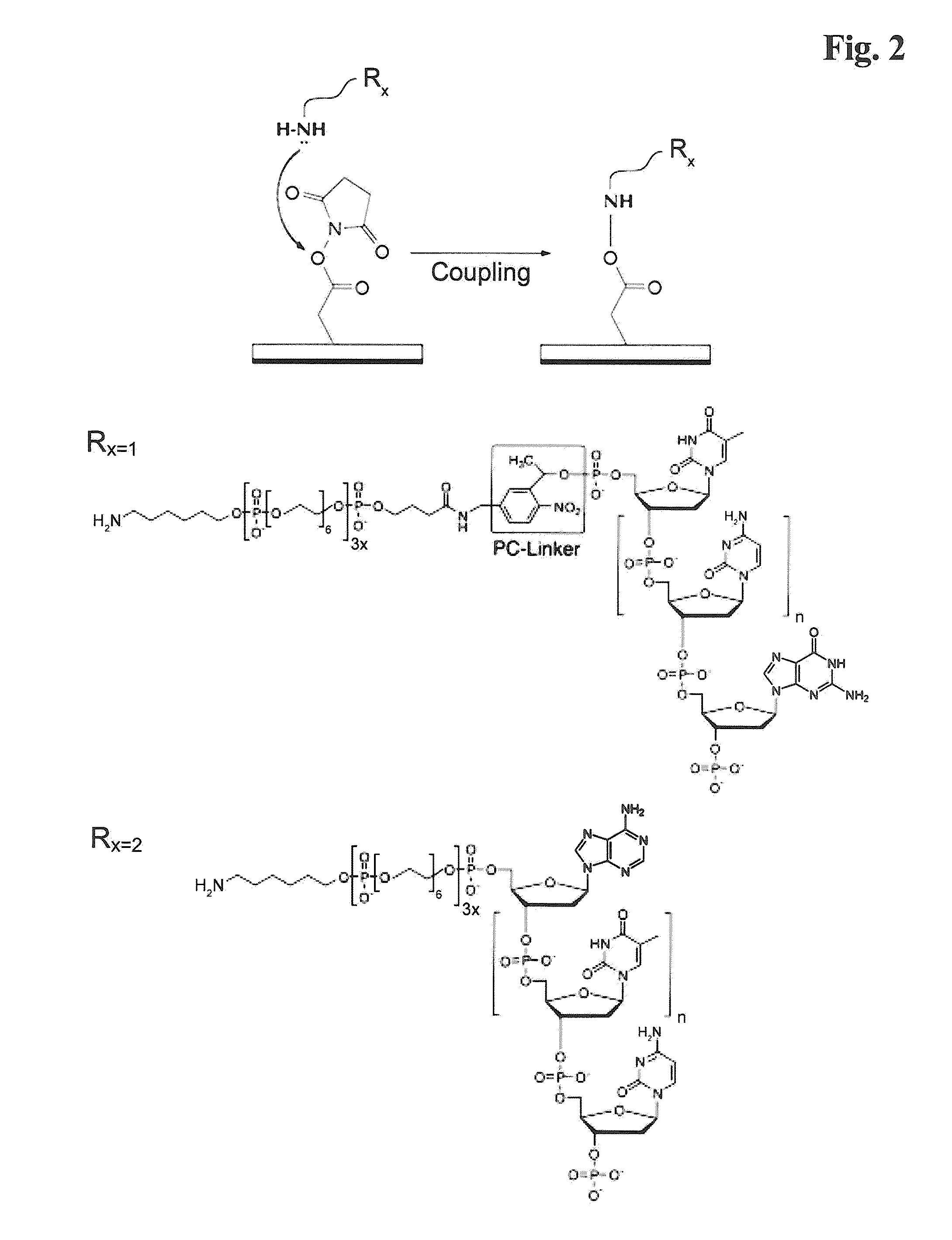
[0205]Amino-modified oligonucleotides containing stationary and photo-cleavable linker (sequence-ID #1-9) respectively, were conjugated to N-hydroxysuccinimide ester (NHS) functionalized sepharose beads (Roche / 454-Life Sciences, Branford, Conn., USA) according to the standard protocol. The chemical reaction mechanism is represented in FIG. 2.
[0206]To trigger photocleavage of the Nitrobenzyl linker these beads were subjected to UV irradiation in a QS1.000 quartz cell (1-cm path length) using a 8 W dual wavelength UV lamp (Camag, Berlin, Germany) at 366 nm. The distance between quartz cell and UV lamp was 2 cm.
example 3
Analysis of the Photocleavage Reaction (Corresponding to FIG. 3)
[0207]This example describes the detection of the photocleavage of fluorescein-modified oligonucleotide probes immobilized to sepharose beads.
[0208]5×106 of beads conjugated with fluorescein-modified oligonucleotide probes (sequence-ID #1) were extensively washed, then suspended in 100 μl 50 mM Tris / HCl pH 7.5 buffer and irradiated for 15 min as described in example 2. Subsequently the beads were centrifuged and the supernatant was analyzed for photocleaved fluorescein-modified oligonucleotides (FAM) by absorbance measurement using a Nanoprop 1000 spectrophotometer (Thermo Scientific, Waltham, Mass., USA) (FIG. 3A).
[0209]In addition, 0.6×106 beads carrying fluorescein-modified oligonucleotide probes (sequence-ID #1) were suspended in 800 μl PCR-reaction mixture (Table 3).
TABLE 3ReagentFinal concentrationTween-800.01%BSA0.10%MgSO42.5mMGlycerol 3.6%Tris-H2SO4 pH 8.958.1mMNH4—SO417.4mMdATP, dGTP, dCTP, dTTPeach 0.40mMExpan...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com