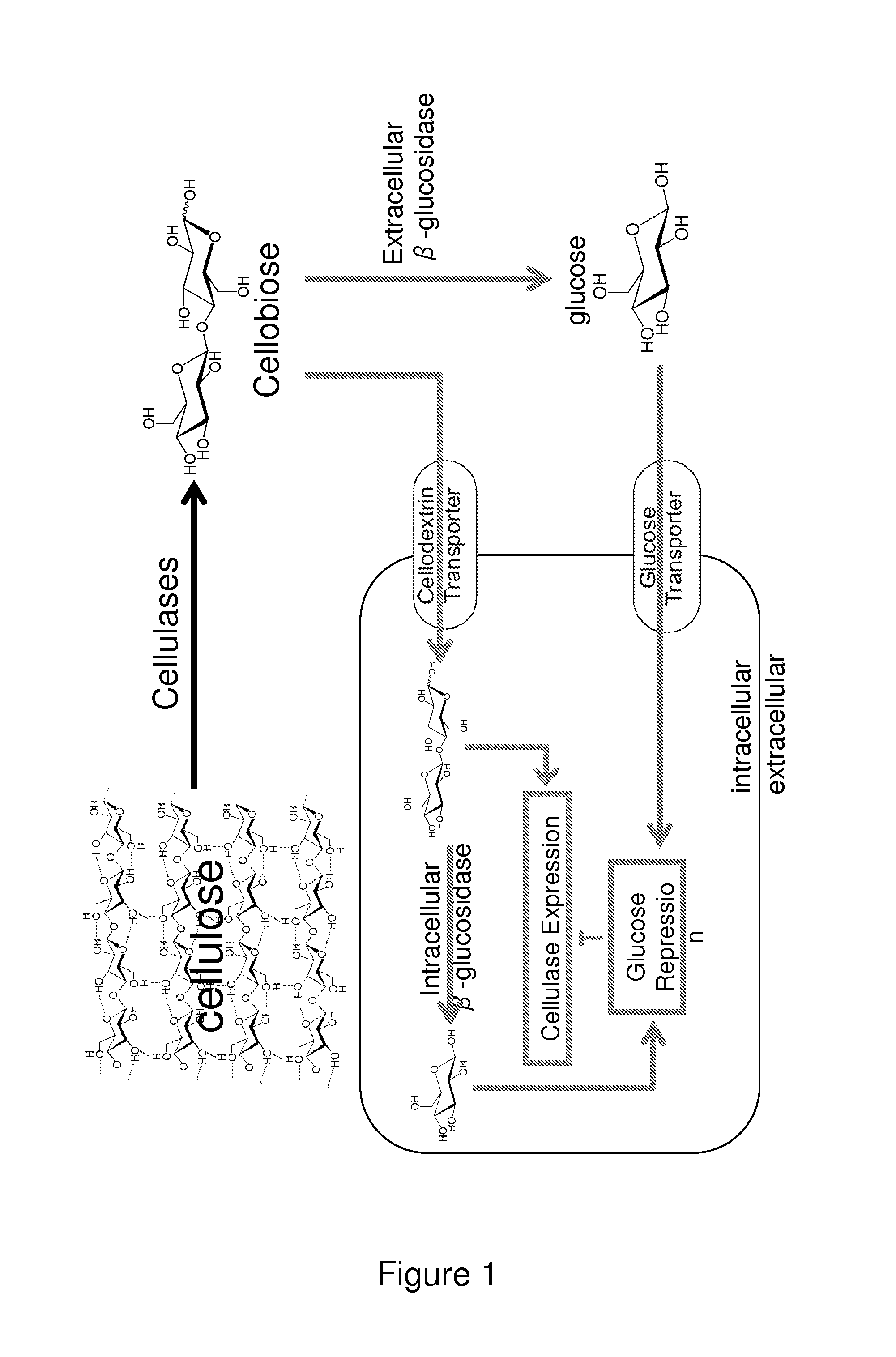
Mutant cells for protein secretion and lignocellulose degradation
a technology of lignocellulose and protein, applied in the field of protein secretion and lignocellulose degradation, can solve the problems of significant technical challenge, major bottleneck in the biofuel production process, and less effective soluble inducers, and achieve the effect of reducing expression, reducing expression, and increasing the expression of cre-1 genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
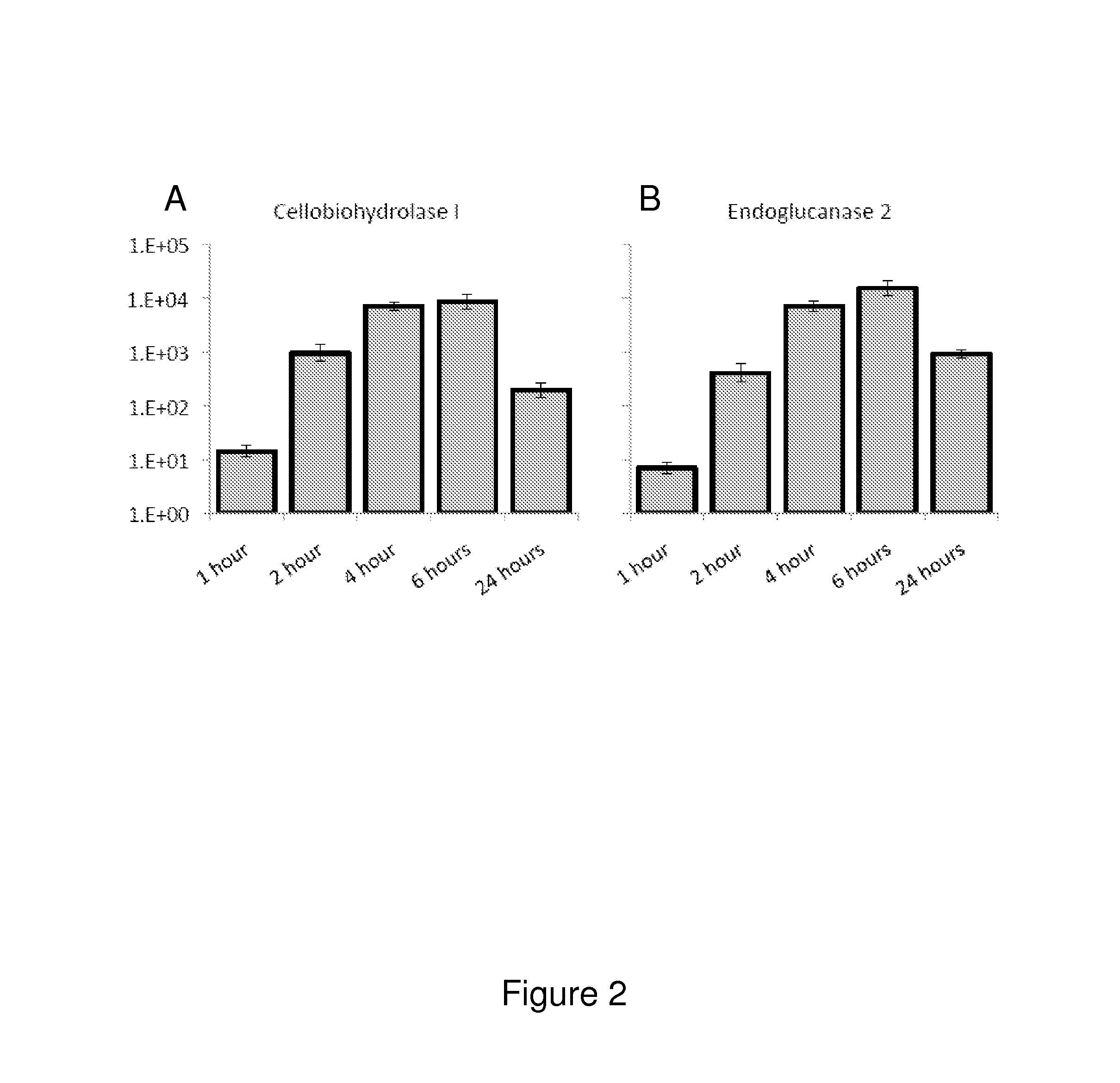
[0145]The following example relates to the characterization of the cellulase transcription and cellulolytic enzyme production induced in the N. crassa triple β-glucosidase gene deletion strain and the triple β-glucosidase and cre-1 gene deletion strain.
[0146]Materials and Methods
[0147]Strains
[0148]Strains were obtained from the Fungal Genetics Stock Center (FGSC) including the Neurospora crassa wild-type (WT) (FGSC 2489), the cre-1 gene deletion (Δcre-1) (FGSC 10372), and deletion strains for the intracellular β-glucosidase NCU00130 (FGSC 11822 and FGSC 11823), and extracellular β-glucosidases: NCU08755 (FGSC 18387 and FGSC 18388) and NCU04952 (FGSC 13731 and FGSC 13732).
[0149]The quadruple deletion produced by performing sequential crosses of the single deletions using the method described by the FGSC (http: / / www.fgsc.net / neurosporaprotocols / How%20to%20make%20a%20cross-2.pdf). The genotype of all deletion strains was confirmed by using a gene-specific primer and a common primer for...
example 2
[0178]The following example relates to the identification of orthologues of the N. crassa β-glucosidase genes NCU00130, NCU04952, and NCU08755.
Materials and Methods
[0179]BLASTp searches were conducted using the National Center for Biotechnology Information (NCBI) non-redundant amino acid database using the NCU00130, NCU04952, and NCU08755 amino acid sequences as queries. Sequence hits from the BLASTp searches were aligned in MEGA5 using ClustalW2.
[0180]Phylogenetic trees were generated using the Neighbor-Joining method (Saitou N. and Nei M., 1987). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl E. and Pauling L., 1965) and are in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted in MEGA5 (Tamura K., Dudley J., Nei M., and Kumar S., 2007).
Results
[0181]The results of the ClustalW amino acid sequence alignments for NCU00130, NCU04952, and NCU08755 orthologues in closely related fungi are shown i...
example 3
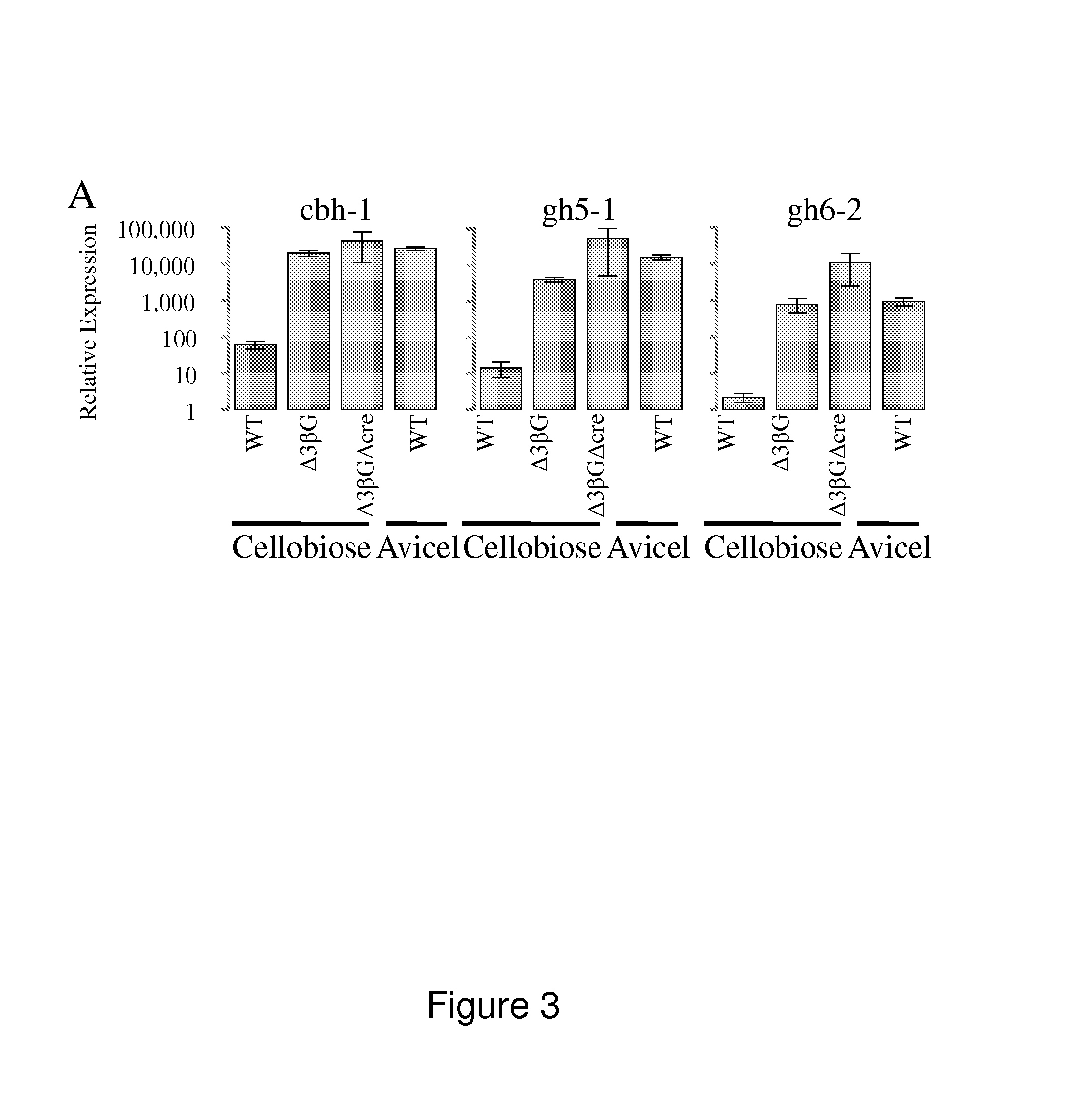
[0185]The following example relates to the identification and characterization of the proteins secreted at higher levels from the triple β-glucosidase gene deletion N. crassa strain, and the triple β-glucosidase and cre-1 N. crassa deletion strain.
Materials and Methods
[0186]Strains
[0187]Strains obtained from the Fungal Genetics Stock Center (FGSC) include the wild type (FGSC 2489), and deletion strains for the intracellular β-glucosidase NCU00130 (FGSC 11822 and FGSC 11823), and extracellular β-glucosidases: NCU08755 (FGSC 18387 and FGSC 18388) and NCU04952 (FGSC 13731 and FGSC 13732). The homokaryon cre-1 deletion strain (NCU08807) is described in (44). Multiple deletion strains were made by performing sequential crosses. The genotype of each multiple deletion strain was confirmed using a gene-specific primer and a common primer for the hygromycin (hph) cassette. The hph forward primer used was SEQ ID NO: 4 from Example 1. The reverser primers used for NCU00130, NCU004953, NCU08755...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com