Methods for nucleic acid capture
a nucleic acid and capture method technology, applied in the field of molecular biology, to achieve the effect of improving quality and yield, fast, reliable and efficien
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
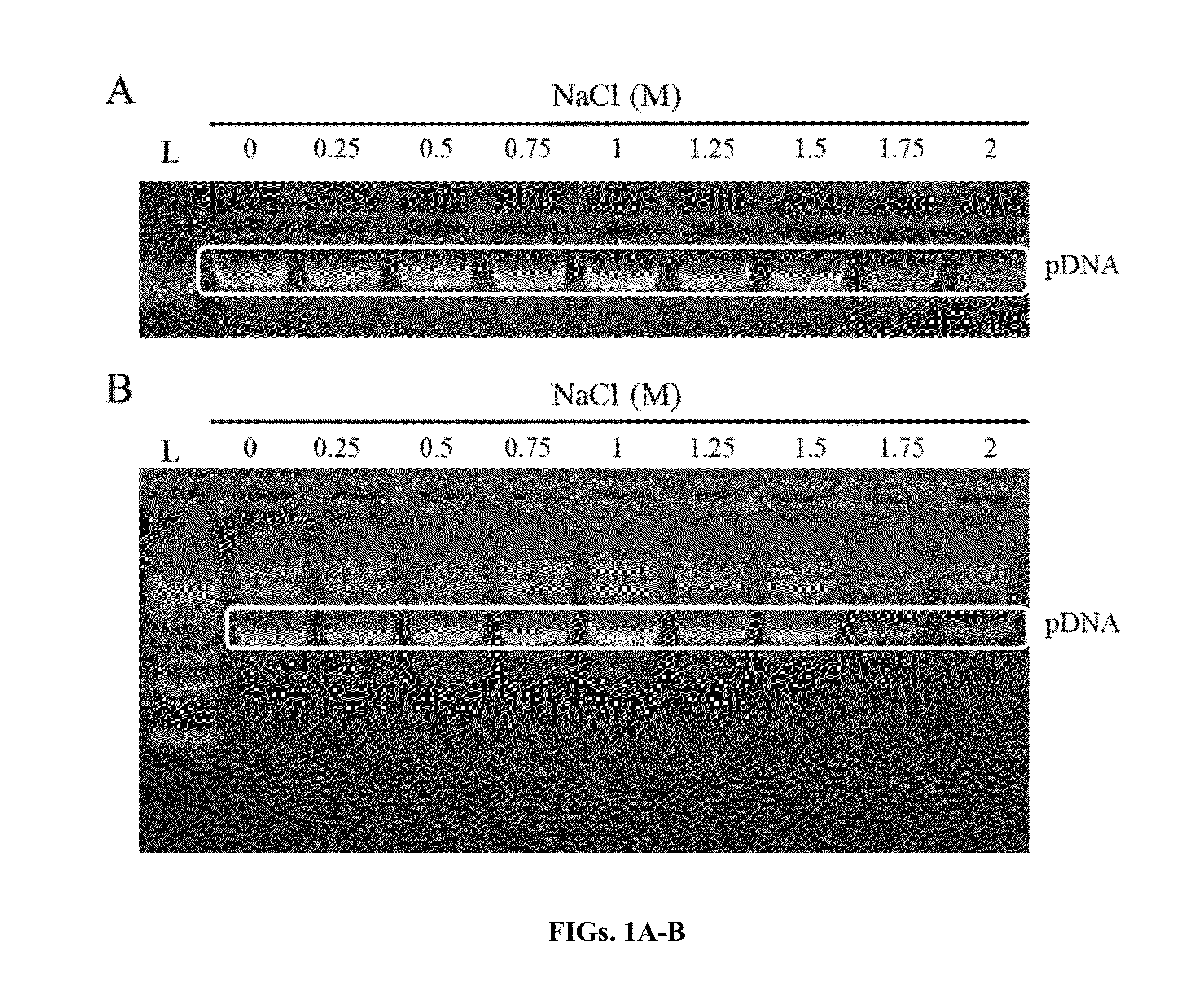
Titration of NaCl with 1-decyl-3-methylimidazolium chloride (DMIC)
[0132]A solution of plasmid DNA (pDNA) and degraded RNA was used to test the ability of 1-decyl-3-methylimidazolium chloride (DMIC) to selectively bind DNA in the presence of varying concentrations of salt. The plasmid DNA used was pGEM, which is a 3.2 kb plasmid. The degraded DNA used was RNA digested with RNAse A, which runs as a smear below the lkb ladder on an agarose gel.
[0133]A TE solution (50 mM Tris-HCl pH 8.0 / 10 mM EDTA) containing pDNA and RNAse A degraded RNA was mixed with P4. The P4 solution (1% DMIC with titration of 0-2M NaCl) was added to the aqueous solution containing the pDNA and degraded RNA and mixed thoroughly by inversion (1 ml of P4 was added for every 3 ml of sample volume). The sample was subsequently loaded onto a glass fiber matrix in a spin column, and the captured nucleic acids were washed with 700 μl 0.5 M NaCl / 80 mM Tris pH 8.5 / 0.5 mM EDTA and subsequently washed with 700 μl 95% ethanol...
example 2
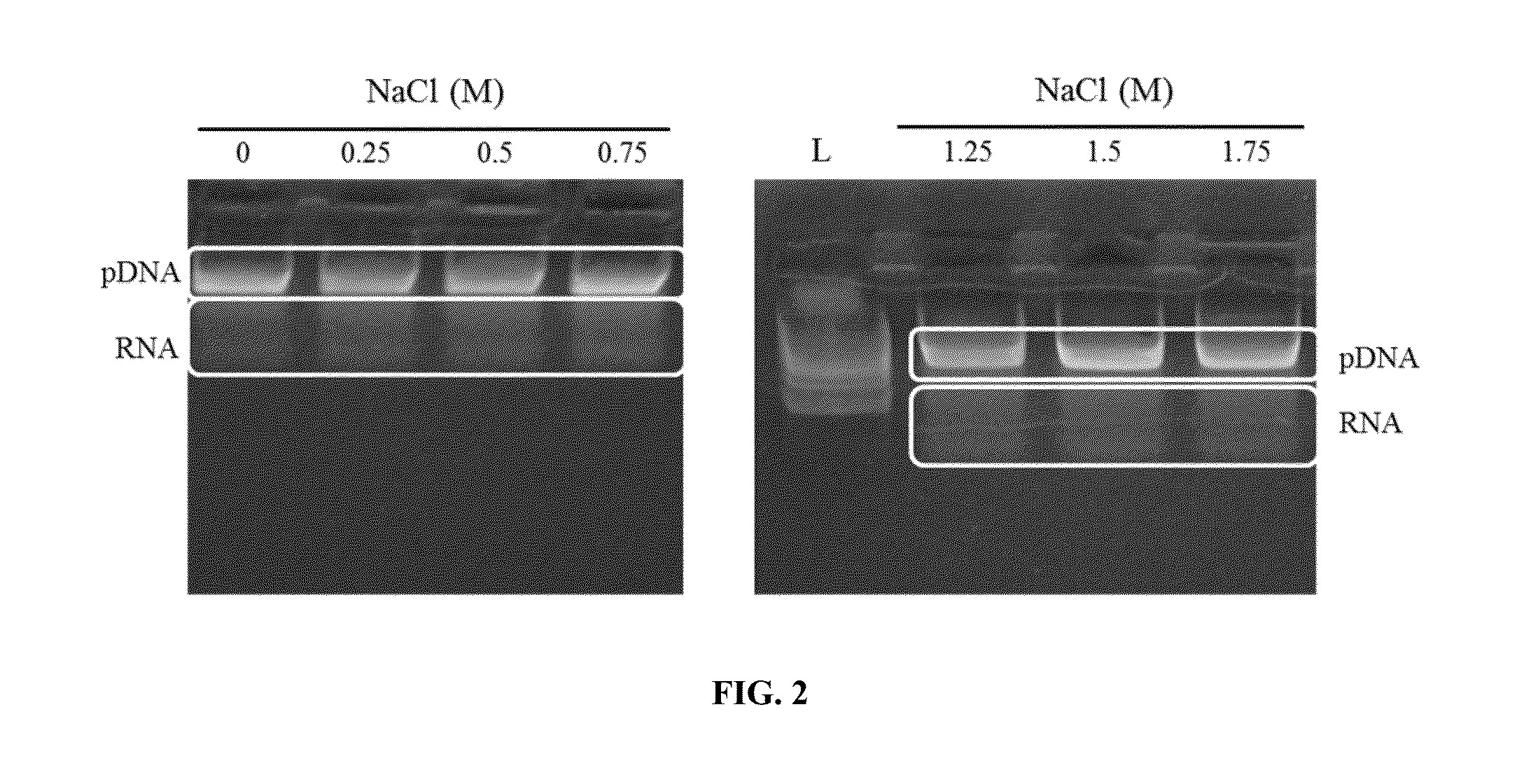
Titration of NaCl with 1,3-didecyl-2-methylimidazolium chloride (DDMIC)
[0134]A solution of plasmid DNA (pDNA) and degraded RNA was used to test the ability of 1,3-didecyl-2-methylimidazolium chloride (DDMIC) to selectively bind DNA in the presence of varying concentrations of salt. The plasmid DNA used was pGEM, which is a 3.2 kb plasmid. The degraded DNA used was RNA digested with RNAse A, which runs as a smear below the 1 kb ladder on an agarose gel.
[0135]A TE solution (50 mM Tris-HCl pH 8.0 / 10 mM EDTA) containing pDNA and RNAse A degraded RNA was mixed with P4. The P4 solution (1% DDMIC with titration of 0-2M NaCl) was added to the aqueous solution containing the pDNA and degraded RNA and mixed thoroughly by inversion. (1 ml of P4 was added for every 3 ml of sample volume.) Next, P4 buffer (1% didecylimidazole with titration of 0-2 M NaCl) was added to the cleared solution and mixed thoroughly by inversion. After loading the solution onto a glass fiber matrix in a spin column, th...
example 3
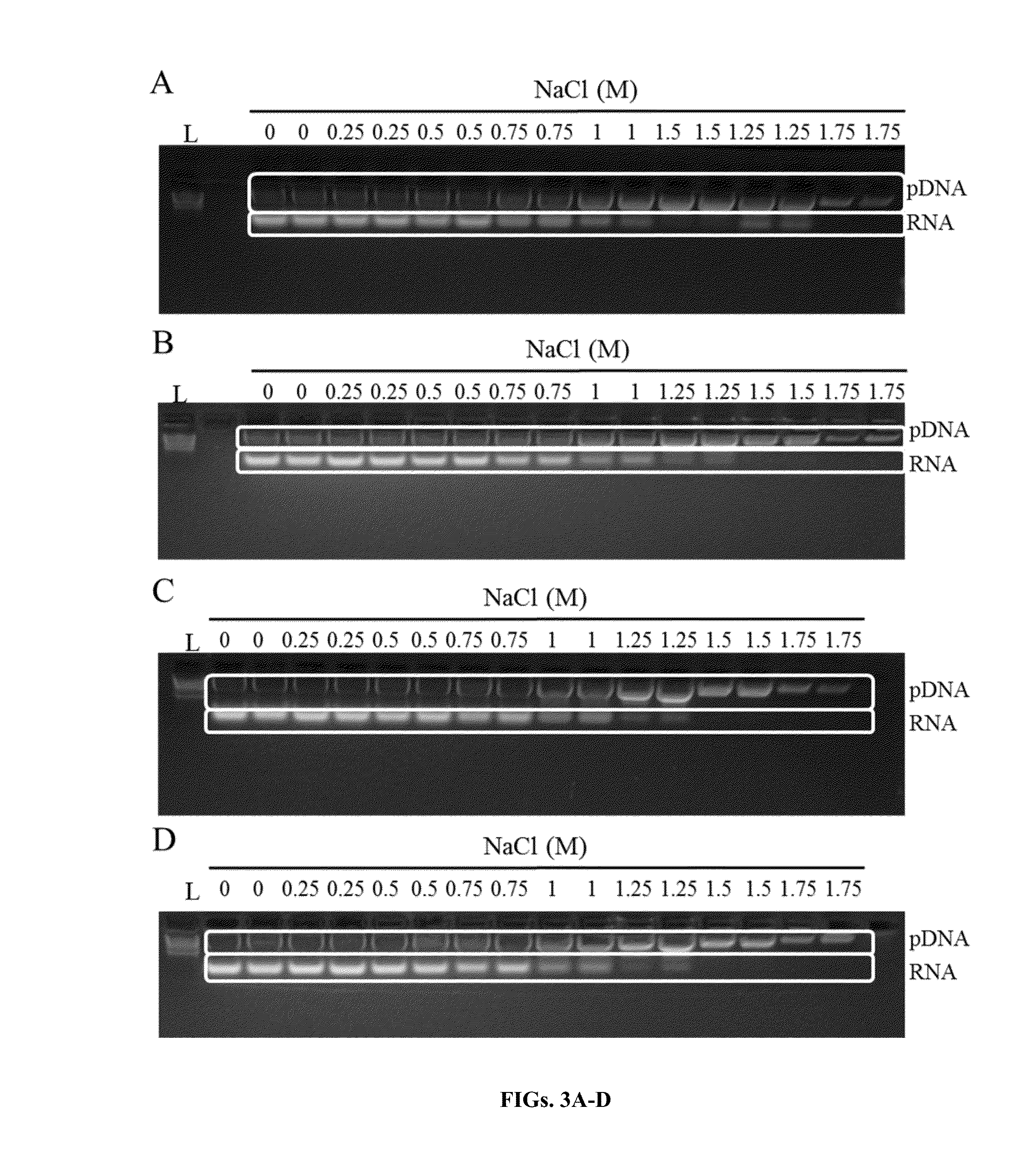
Titration of NaCl with cetylpyridinium bromide (CPB)
[0137]A solution of Cetylpyridinium bromide (CPB) was evaluated with increasing concentrations of sodium chloride to determine the optimal concentrations of the salt and phase separation reagent for selective capture of plasmid DNA while removing degraded RNA. For this, a culture containing JM109 transformed with pGEM was grown overnight for 16 hours. The cells contained in 35 ml of culture were pelleted and suspended in 5 ml P1 buffer. The cells were then lysed using 5 ml P2 buffer for 3 minutes and the solution was neutralized and genomic DNA / proteins were precipitated using 5ml P3 buffer that contained 1.5 M potassium acetate at about a pH 4.9. The precipitated cellular debris were cleared by using a glass fiber filter. 5 ml P4 buffer (0.25%-1% CPB with titration of 0-1.75 M NaCl) was added prior to loading the solution onto a glass fiber matrix in a spin column. The matrix was washed with 700 μl 0.5 M NaCl / 80 mM Tris pH 8.5 / 0.5...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com