Nanostructured separation and analysis devices for biological membranes
a biological membrane and nanostructure technology, applied in the field of nanostructured matrices fabrication, can solve the problems of lack of reusability, set of separation strategies that rely on this technique, and difficulty in incorporation of these techniques
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
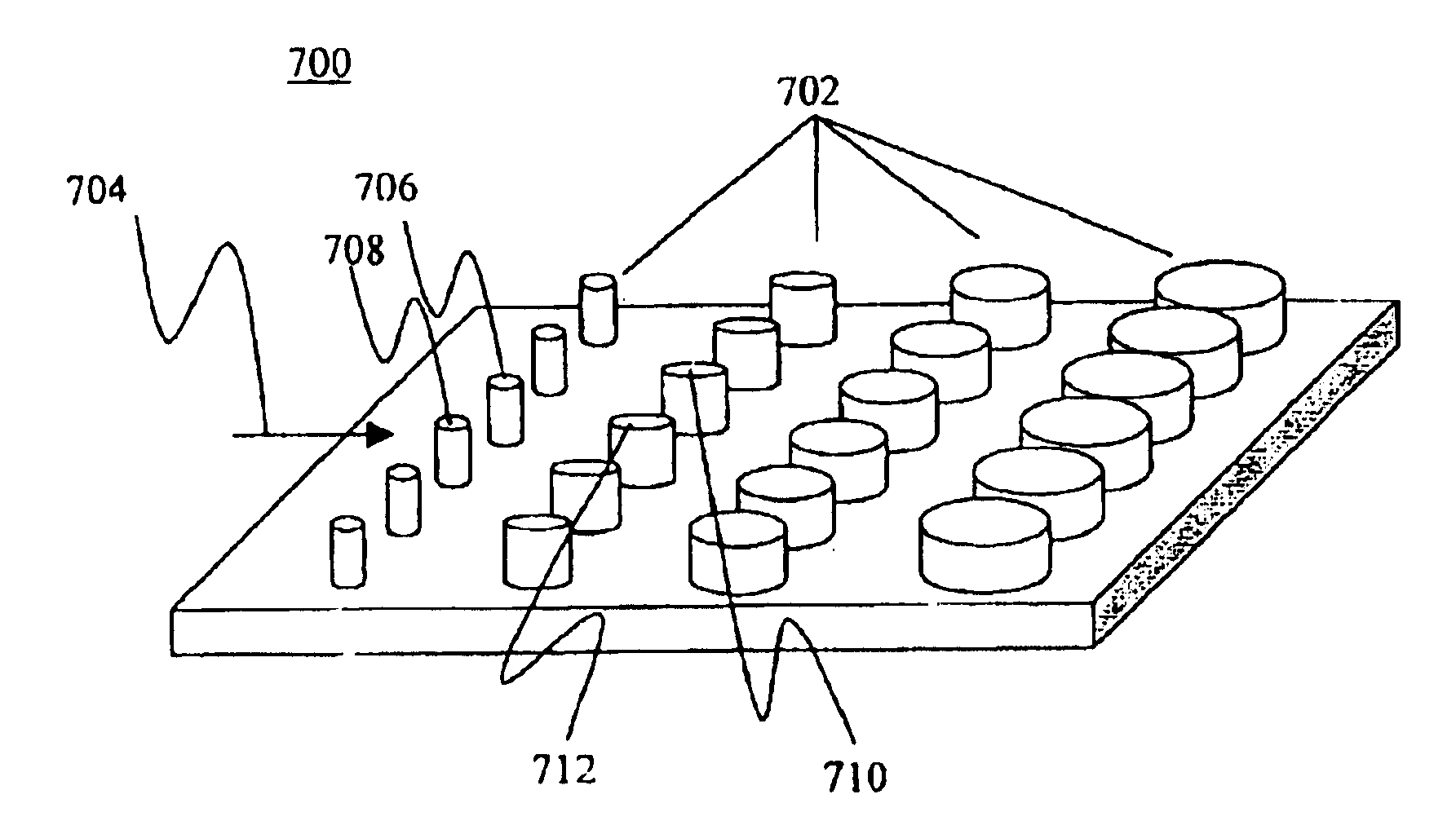
[0133]Design and construction of microscale electrophoresis cells incorporated much of the characteristics of the present invention into a compact system. The cell preferably has the following characteristics: (1) electrochemical current and fluid flow is restricted to occur only through the separation matrix; (2) loading and stacking functions are included; (3) monitoring of mobility and biomolecular detection is possible (e.g., through fluorescence imaging); and (4) for certain applications, separated compounds are recoverable. Simple methods have been used for incorporating nanostructured silicon / silica chips into electrophoresis cells that satisfy criteria (1-3) above. For example, simple methods of rapid prototyping of elastomeric gasket materials have been used.
example 2
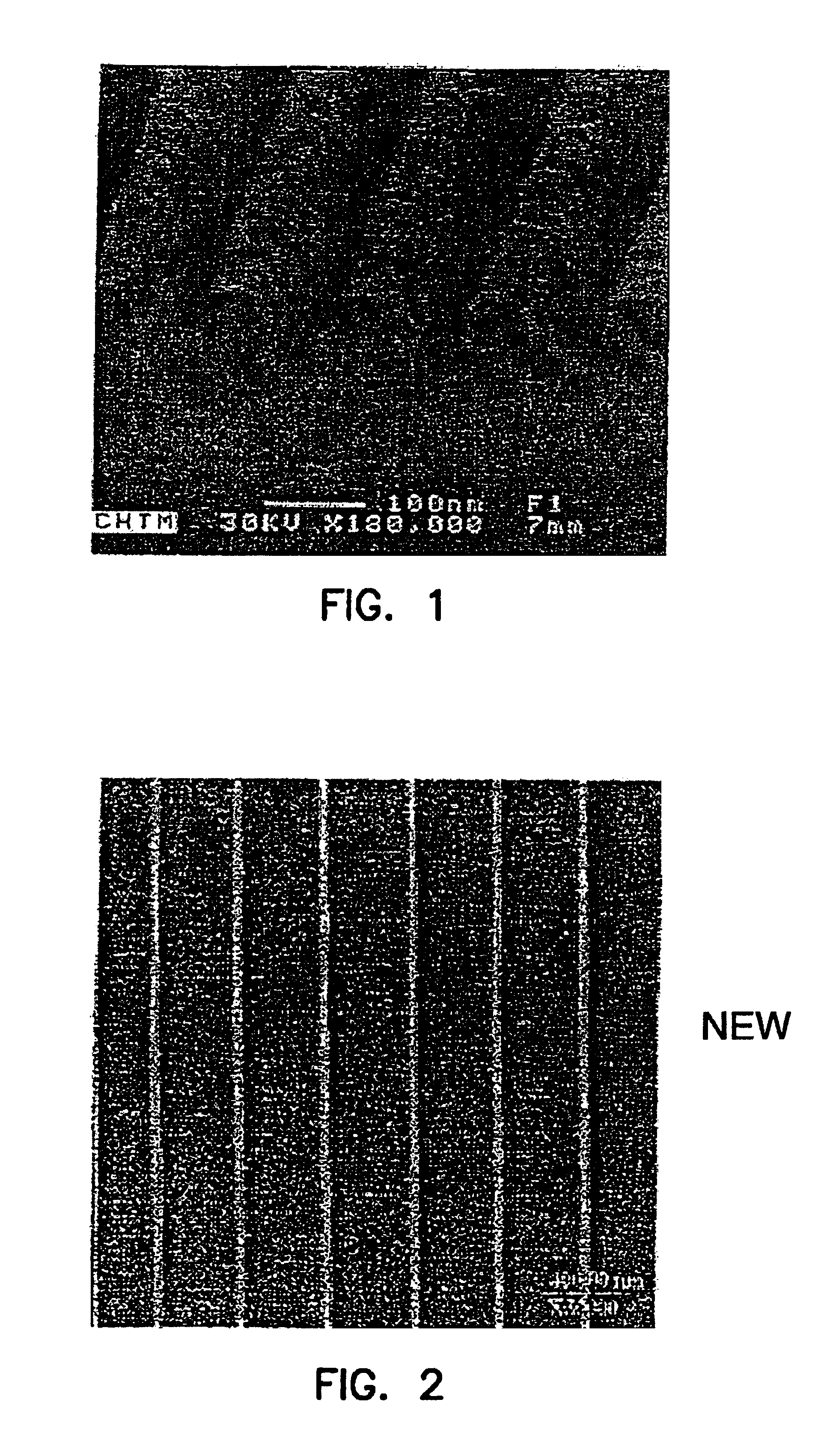
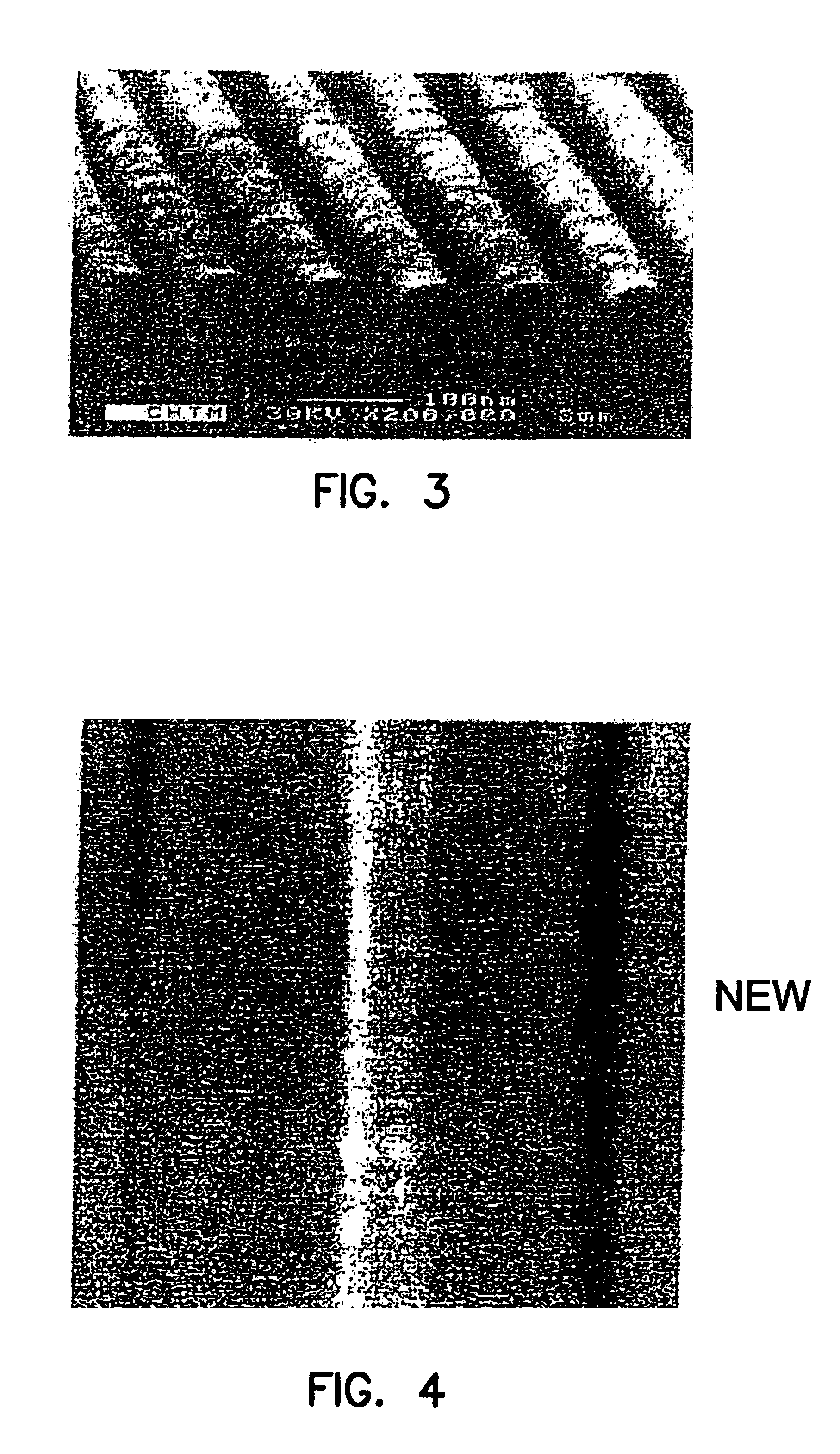
[0134]Supported phospholipid bilayers (SPBs) of egg phosphatidyl choline (Egg PC) were formed by vesicle fusion on nanostructured silicon wafers containing troughs ˜180 nm in width on a 360 nm pitch. An intercalating dye was introduced, and the membranes were imaged by scanning laser microscopy. The resultant fluorescence micrographs indicated that the SPBs formed uniformly over the surface and simple FRAP measurements indicated that the bilayers were fluid and that recovery of fluorescence was preferentially in the direction parallel to the nanotroughs.
example 3
[0135]Transmembrane or membrane associated proteins may be incorporated into an SPB, from incorporation in the vesicle stage, insertion on the membrane or through incorporation of cell ghosts (i.e., intact membranes isolated from cells or organelles). The architecture and / or chemistry of the underlying nanotextured support would be then used to guide the movement of membrane proteins through the supported or suspended bilayer, either by size exclusion of the trough over which the membrane is supported, or by chemical interactions with modifications on the nanostructured support.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
| refractive index | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com