Yeast expression high specic activity phytase gene obtained using chemical synthesis and molecular evolution
A technology for synthesizing genes and yeast expression vectors, applied in plant gene improvement, biochemical equipment and methods, genetic engineering, etc., can solve the problems of low expression of original strains, easy degradation of mutagenized strains, and high production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1: the chemical synthesis of acid phytase gene
[0027]1. The thermostable phytase gene was synthesized by continuous extension PCR. The length of the primer is 70-90bp, 20 oligonucleotide primers are synthesized, the primers are connected by 20-30bp overlapping sequences, and the Tm value is 60-66. Add all primers to the reaction system, the amount of primers in the middle is 10-20ng, and the amount of primers on both sides is 100-200ng. The PCR reaction system is 100 μL. The PCR amplification conditions were 94°C, 30s; 65°C, 30s; 72°C, 2min. A total of 35 cycles were performed.
[0028] Acid phytase gene synthesis primers:
[0029] phyi1:
[0030] TTGGCTGTCCCAGGCTTCCAGAAACCAGTCCTCTTGTGACACTGTTGATCAAGGTTATCAATGCTTCTCCGAG
[0031] phyi2:
[0032] GTTATCAATGCTTCTCCGAGACTTCTCACTTGTGGGGTCAATACGCTCCATTCTTTTCCTTGGCTAACGAATCCGTCATTCTCTCCAGAAG
[0033] Phyi3:
[0034] GAATCCGTCATTCTCTCCAGAAGTTCCAGCTGGTTGCAGAGTCACCTTCGCTCAGGTCTTGTCCAGACATGGTGCTAGATACCCAACTGA...
Embodiment 2
[0069] Embodiment 2: the DNA molecular rearrangement (DNA Shuffling) of acid phytase gene
[0070] 2.1PCR amplification of acid phytase gene and recovery
[0071] Using the synthetic phytase gene as a template, phyiZ1 and phyiF1 as primers to amplify the phytase gene, phyiZ1: (5'-TTGGATCCTTGGCTGTCCCAGCTTCCAGAAAC-3'); phyiF1: (5'-CGAG CTCTTAAGCGAAGCATTCAGCCCAGTCAC-3') The reaction conditions are: 94 Pre-denaturation at ℃ for 10min, denaturation at 94℃ for 30s, annealing and extension at 72℃ for 1.5min, a total of 30 cycles, 1% Agrose electrophoresis, and recovery of a 1.4kp gene fragment by the suction bag method.
[0072] 2.2 DNase I degrades DNA and recovers small fragments
[0073] Recover the phyi gene fragment with DNase I buffer (50mmol / L Tris-ClpH7.4+1mmol / L MgCl 2 ) in 100 μl; add 0.1U DNase I and treat at 25°C for 15 minutes. 70°C for 10 minutes. 10% acrylamide electrophoresis, recovery of small fragments of 10-50bp by suction bag method. The precipitate was disso...
Embodiment 4
[0082] Example 4: Yeast expression phytase vector construction
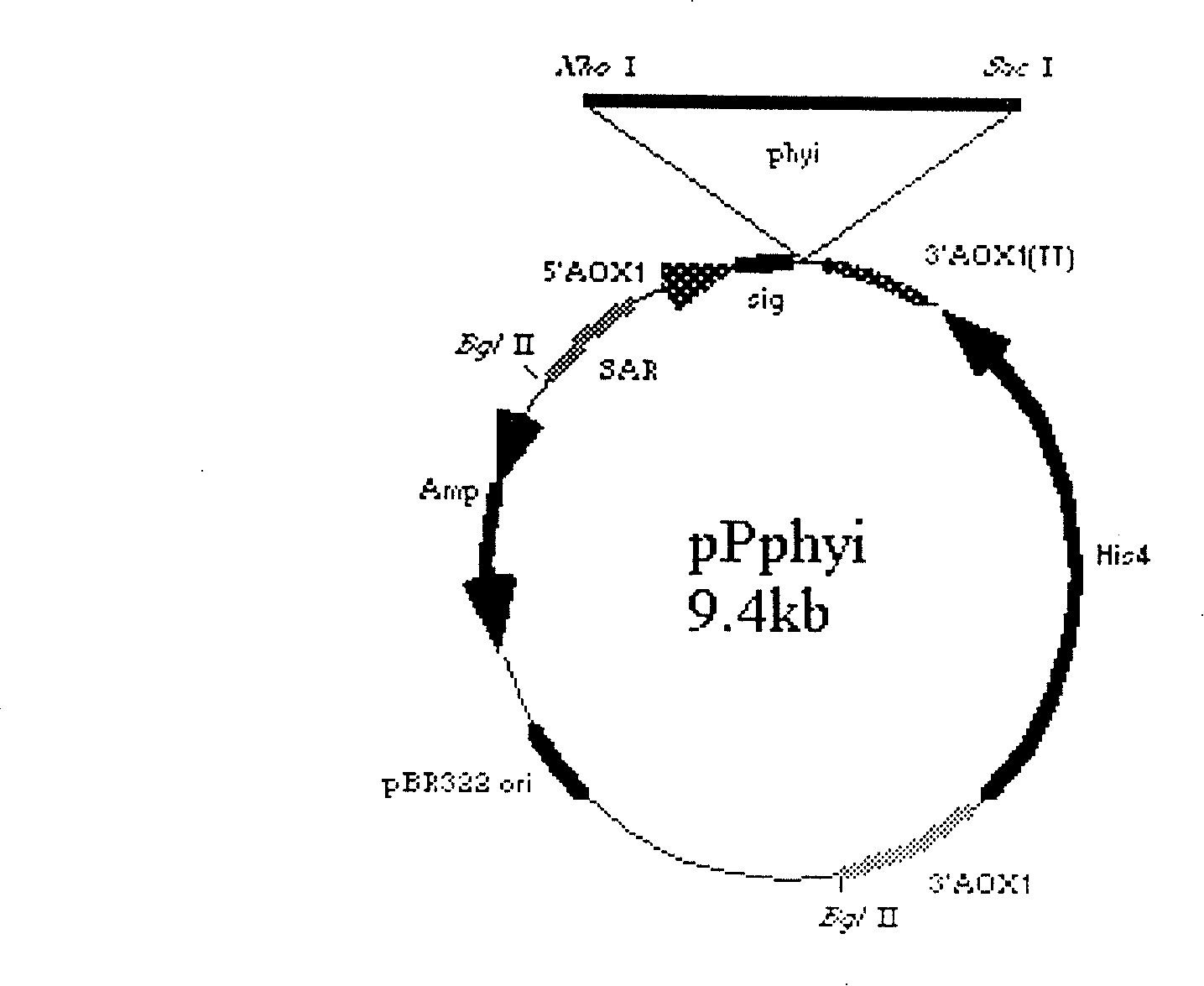
[0083] Design primers according to the sequences at both ends of the synthetic gene, add Xho I cutting point and signal state cutting sequence at the 5' end of the gene, the primer is: PHY1Z (5'-AACTCGAGAAAAGAGAACCTCCG GATTGGCTGTCCCAGGCTTCCAG AAACCAGTCC-3'), add Not I at the 3' end of the gene Cutting point: the primer is: PHY1F (5'-AACG CGGCCGCTTAAGCGAAGCATTCAGCCCAGTC ACCACCGGTAC-3'). After the amplified fragment was cloned, it was digested with Xho I and Not I, inserted into the pPIC9 vector (Invitrogen company product), and constructed into the yeast expression vector pPphyi of phyi ( figure 1 ).
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com