Evolved immunoglobulin binding molecule, and its preparation method and uses
A technology of immunoglobulin and binding molecules, which is applied in biochemical equipment and methods, chemical equipment and methods, botanical equipment and methods, etc., can solve the problems of LD5 functional characteristics without further elaboration, and achieve improved sensitivity and accuracy , low cost, good effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0139] Example 1 A method for prokaryotic preparation of a novel evolutionary immunoglobulin binding molecule
[0140] 1. cDNA sequence and cloning of new evolved immunoglobulin binding molecules
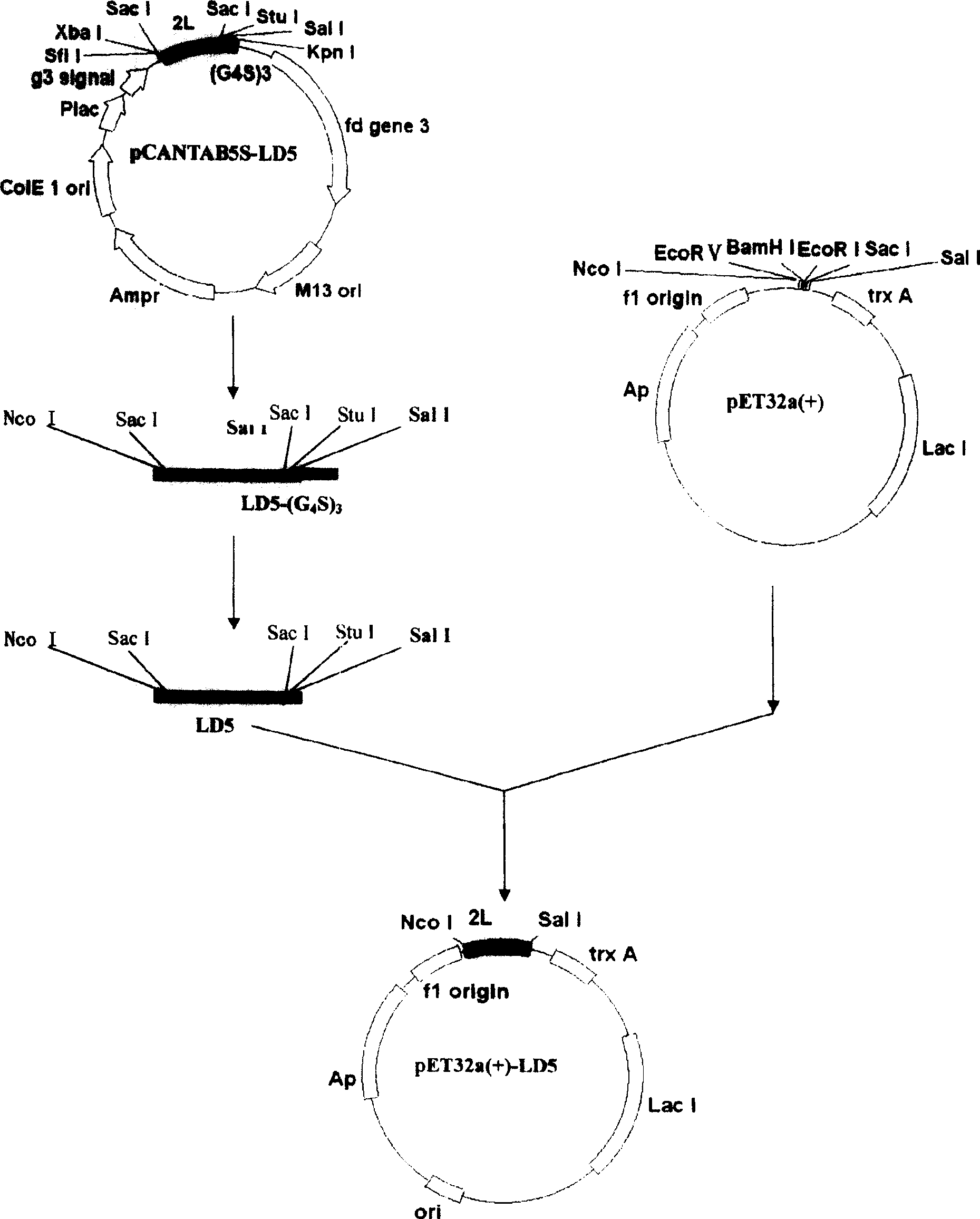
[0141] Recombinant phagemid vector pCANTAB5S-LD5, cloned with cDNA sequence encoding evolutionary immunoglobulin binding molecule LD5 (LDLDL; L is the B3 domain of PpL, D is the D domain of SpA), was obtained through molecular evolution screening and preparation The specific steps of pCANTAB5S-LD5 are described in "Phage Display SpA and PpL lg Binding Single Domain Random Combinatorial Library and Affinity Screening", Progress in Biophysics and Biochemistry, 2005 Vol. 32, No. 6, pages 535-543 Disclosure, that is, the 27# / 44# in the third round of screening and the 4# / 33# cloning vector in the fourth round of screening in Table 4 in the literature. The cDNA sequence is shown in SEQ ID No. 1, and the amino acid sequence is shown in SEQ ID No. .2 shown.
[0142] Evolutionary immunoglobulin...
Embodiment 2
[0174] Example 2 Binding of novel evolutionary immunoglobulin binding molecules to various antibodies and antibody fragments
[0175] 1. Combination with human polyclonal IgG:
[0176] 1.1 Human polyclonal IgG was purchased from Sigma, and PpL and SpA were purchased from Sigma.
[0177] 1.2 1mg / ml human polyclonal IgG was dialyzed with PBS (pH 7.2). Take 50μL of 3mg / ml long-arm activated biotin (purchased from PIERCE) and add 1mL IgG (1mg / mL). After shaking slightly at room temperature for 4h, add a dialysis bag and dialyze in PBS overnight at 4℃. After dialysis, add the same amount of glycerol to Store at -20°C.
[0178]1.3 After adjusting the concentration of LD5, PpL and SpA to 1mg / ml, the 96-well plate was coated with carbonate buffer (pH9.6) 1:200, and each protein was coated with 3 rows of multiple wells. After 24h at 4℃ Wash the plate with PBST for 4-5 times, then wash the plate with blocking solution (2% BSA 0.05% TWEEN-20) at 37°C for 1 hour. The biotin-labeled IgG starts...
Embodiment 3
[0201] Example 3 Confirmation of the dual-site binding mode of the new evolved immunoglobulin binding molecule κ light chain and VH3 heavy chain
[0202] SpA has a strong affinity with the Fc segment of IgG, and SpA can also bind to the VH region belonging to the VH3 gene family (SassoEH, Silvcrman GJ, Mannik M. Human 1gA and IgG F(ab')2 that bind to staphylococcalSpA belong to the VHIII subgroup. J Immunol. 1991:147:1877-1883). PpL binds to immunoglobulin by interacting with the light chain of Ig, mainly subtypes 1, 3, and 4 of the kappa light chain (NilSon BH, Solomon A, BjorckL, et al. PpL from peptostreptococcus magnus binds to the kappa light chain variabledomain; J Biol Chem. 1992; 267(4): 2234-9). From the binding experiments of LD5 and various Ig molecules, LD5 has a higher affinity for IgGFab IgM IgA than SpA and PpL. It is speculated that LD5 has dual-site binding properties for the variable regions of VH3 heavy chain and kappa light chain. The competitive inhibition tes...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com