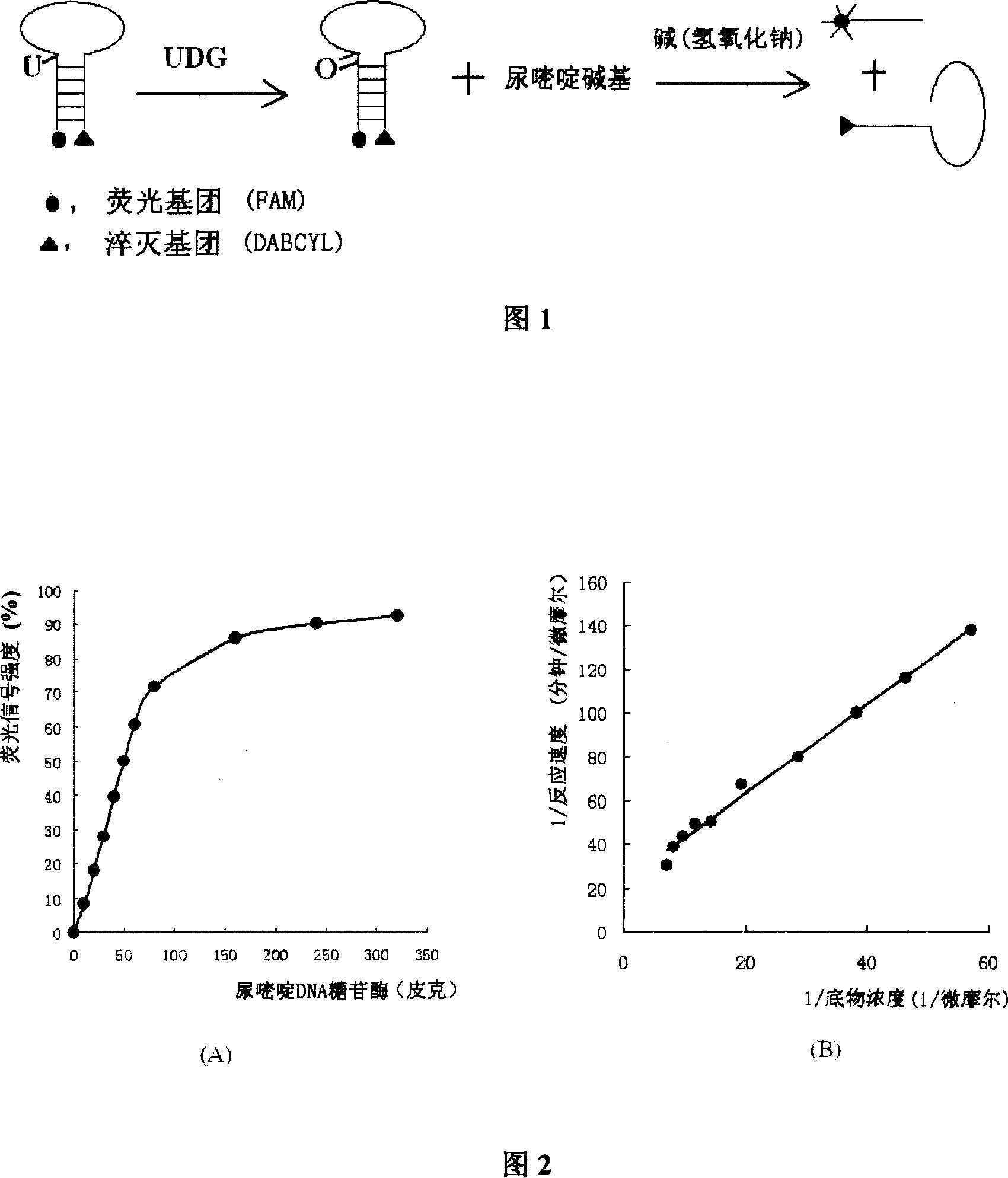
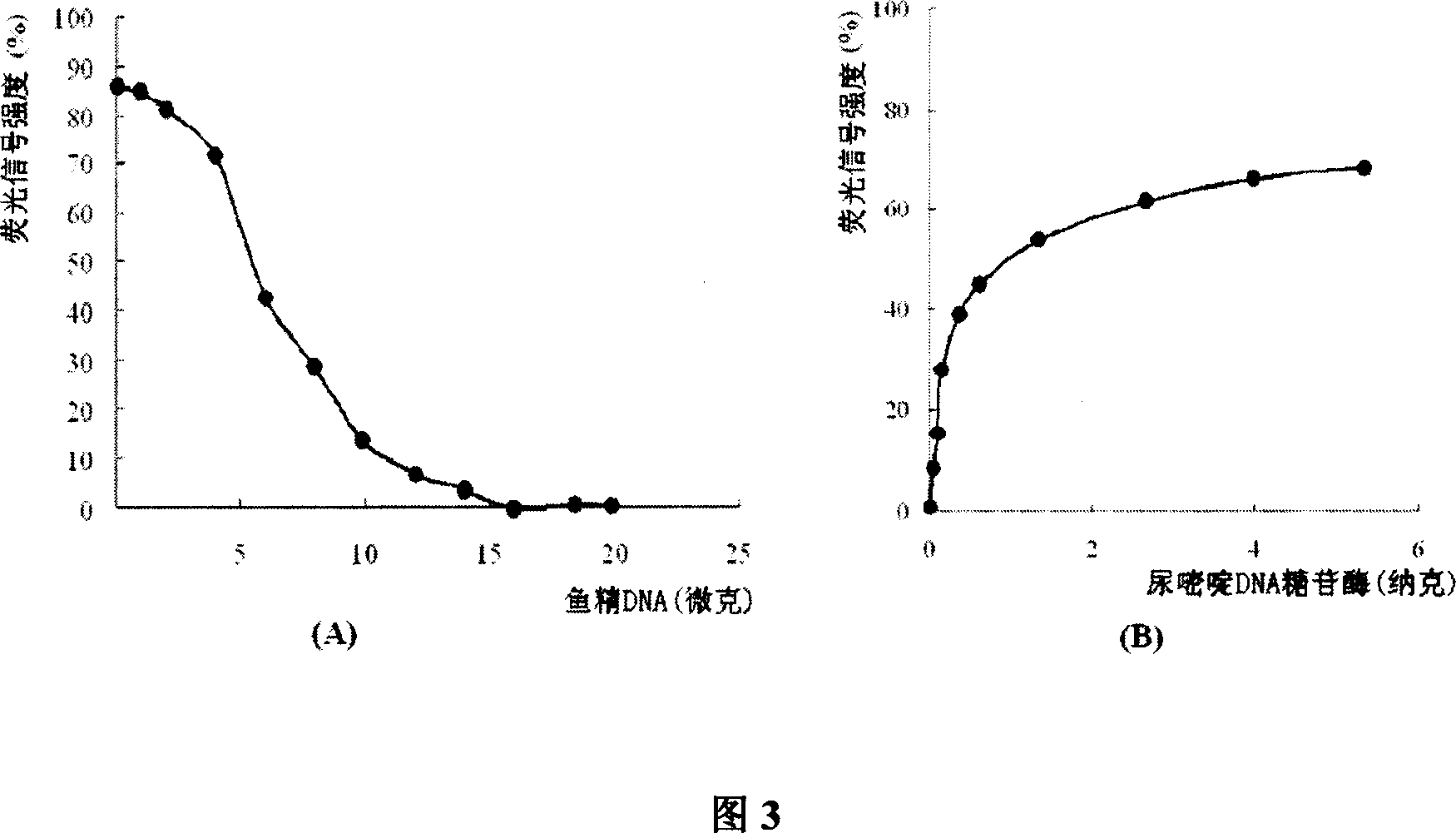
Method for detecting ura DNA glycosidase activity adopting molecular beacon as substrate
A molecular beacon and uracil technology, applied in the field of protein qualitative and quantitative detection, can solve the problems of cumbersome operation process and low sensitivity, and achieve the effect of reducing damage, saving experimental time, and safe UDG activity detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1 pure UDG activity detection
[0027] 1. Design and synthesis of molecular beacons:
[0028] Refer to the oligonucleotide substrates used to detect UDG activity in the past, and then determine the primary sequence of the molecular beacon according to the principle of the lowest energy. DABCYL marker, and 7 pairs of sequences are complementary pairs to form a stem structure, such as 5'(7-FAM)-GACCTGC and GCAGGTC-(DABCYL)3'. The molecular beacon sequence is 5'(6-FAM)-GACCTGCUCAGTACGAGAGGACCGCAGGTC-3'
[0029] 2. Preparation of tested UDG samples:
[0030] Escherichia coli BL21(DE3) carrying the protein expression plasmid of Pdest17-UDG was grown at 37 degrees in LB medium containing 50ug / ul Kanade. When the OD600 value was 0.8, IPTG (isopropylthio-β-D-galactoside) was added to a concentration of 1Mm for induction. After 6 hours of induction, the cells were collected and lysed by adding lysate. The lysate was then centrifuged at 12,000 rpm for 45 minutes, ...
Embodiment 2
[0037] The detection of UDG activity in the crude extract of embodiment 2
[0038] 1. Design and synthesis of molecular beacons:
[0039] Same as Example 1, the substrate is still the above-mentioned molecular beacon.
[0040] 2. Construction of UDG mutant strain and preparation of crude extract:
[0041] a. Design of primers for UDG mutant strains:
[0042] First, design the primers for constructing mutant strains. In the primers, 20nt oligonucleotides are required to match the Pet28A plasmid template without the UDG coding sequence. In addition, 40nt oligonucleotide sequences are required to be compatible with the E.coli DY329 strain The adjacent sequence of the UDG coding gene is homologous. In summary, the primer sequences were designed as follows: Forward primer (5'-ttaagctagg cggattgaag attcgcagga gagcgag atg ggcagcagccatcatcatca tcatca -3') reverse primer (5'-agccgggtgg caactctgcc atccggcatttccccgcaa a tttggtatct gcgctctgct gaagcc ).
[0043] First, the sec-kan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com