DNA encoding novel enzyme having D-serine synthase activity, method of producing the enzyme and method of producing d-serine by using the same
A technology of serine and DNA sequence, applied in the field of preparation of D-serine, can solve the problems such as no
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0129] (Acquisition of the gene encoding DSA)
[0130] Achromobacter xyloseoxidans (ATCC9220), Achromobacter denitrificans (NBRC15125) and Achromobacter xyloseoxidans (NBRC13495) were inoculated in 50 ml of LB medium. After culturing overnight at 30°C, the bacteria were collected and lysed with a lysozyme solution containing 1 mg / ml. After treating the lysate with phenol, the DNA was precipitated by ethanol precipitation using the usual method. The resulting DNA pellet was recovered by winding it on a glass rod, washed, and used for PCR.
[0131] As primers for PCR, oligonucleotides (synthesized on request to Hokkaido System Science Co., Ltd.) having base sequences shown in SEQ ID NOs: 1 and 2 designed based on the known DTA gene were used. The above primers have restriction enzyme recognition sequences of KpnI and HindIII near the 5' end and near the 3' end, respectively.
[0132] Use 0.025ml of PCR reaction solution containing 6ng / μl of the chromosomal DNA of the above-me...
Embodiment 2
[0141] (Preparation method of DSA)
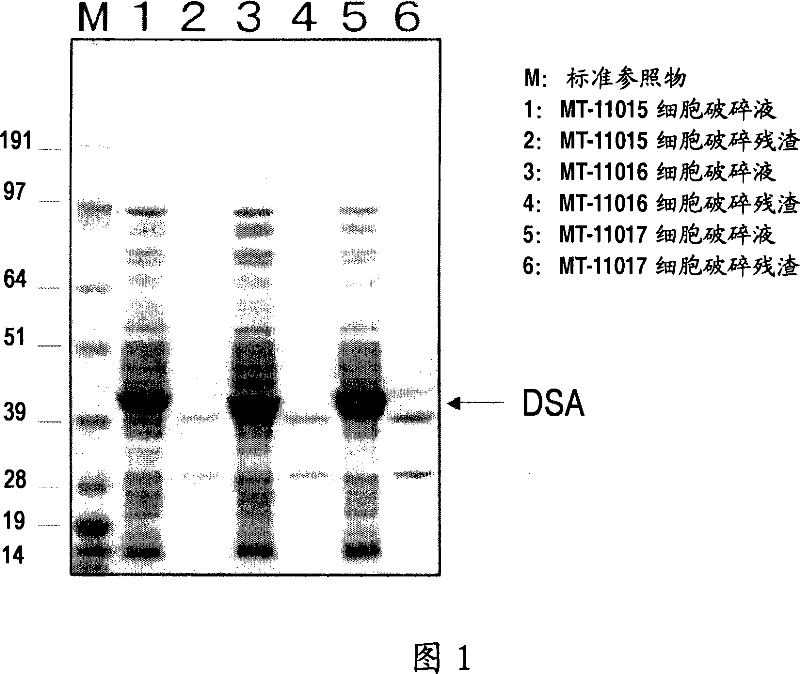
[0142] 0.5 mL of the transformant suspension prepared in Example 1 was disrupted in ice water for 5 minutes using a Bioruptor (manufactured by OLYMPUS). The broken solution of the transformant was centrifuged to prepare the broken solution. The disrupted solution was analyzed by SDS-polyacrylamide electrophoresis, and the results are shown in Figure 1.
[0143] 0.5 mL of 100 mM Tris-HCl buffer (pH 8.0) was added to the pellet, and the same analysis was performed on the cell residue.
[0144] Protein expressed at about 40 kDa was seen in the soluble fraction of all transformants and not in the insoluble fraction. The molecular weight almost coincides with the molecular weight of the amino acid sequence estimated from each gene.
Embodiment 3
[0146] (Purification of DSA and synthesis of D-serine by purification of DSA)
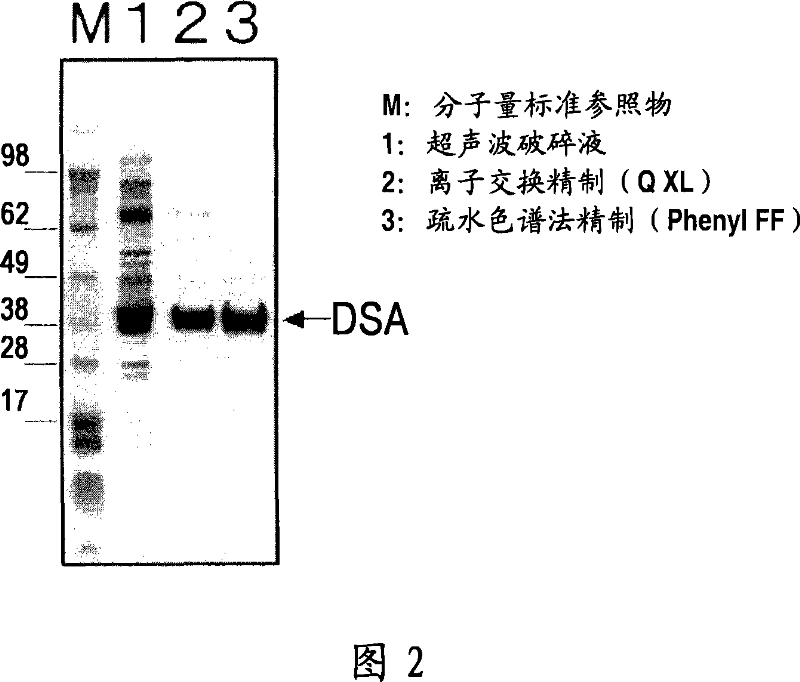
[0147] Centrifuge 10 mL of the disrupted MT-11015 solution prepared by the same method as in Example 2 at 10,000 rpm for 20 minutes to prepare an enzyme solution for removing cell residues. An anion exchange resin (HiTrap Q-XL, manufactured by Amersham) was adsorbed on the enzyme solution, and it was eluted with a linear gradient from 100 mM Tris-HCl buffer (pH 8.0) containing 10 mM magnesium chloride and 50 mM sodium chloride to 100 mM containing 10mM magnesium chloride and 500mM sodium chloride in Tris-HCl buffer (pH8.0). The hydrophobic chromatography resin (HiTrap Phenyl FF, manufactured by Amersham) was allowed to adsorb the active fraction, and it was eluted with a linear gradient from 100 mM Tris-HCl buffer (pH 8.0) containing 10 mM magnesium chloride saturated with ammonium sulfate to 100 mM Tris-HCl buffer (pH 8.0) containing 10 mM Magnesium chloride in Tris-HCl buffer (pH 8.0). It shoul...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com