bzip transcription factor related to corn kernel growth, its coding gene and gene expression
A technology for transcription factors and coding genes, which can be used in genetic engineering, plant genetic improvement, fermentation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Obtaining a new gene of bZIP transcription factor expressed during corn kernel development
[0026] 1. Construction of a homogenized cDNA library at corn kernel development stage
[0027] In order to obtain the expression sequence of the entire grain development stage, collect corn grains 3 to 36 days after pollination, mix them in equal volumes, and extract total RNA (Trizol kit, Qiagen) to obtain the PolyATtract from Promega R The mRNA purified by the mRNA Isolation System III Kit is used as a template, and a cDNA homogenization scheme based on the principle of renaturation kinetics is used, that is, through denaturation and renaturation operations, the abundance of cDNA tends to be consistent, and finally a homogenized cDNA library is established .
[0028] 2. Acquisition of genes encoding bZIP transcription factors
[0029]The sequencing universal primer T3 / T7, dideoxy method (DYEnamic ET-Terminator Cycle Sequencing), and DNA sequence analyzer (MegaBACE 4500)...
Embodiment 2
[0030] Example 2: Identification of bZIP transcription factors
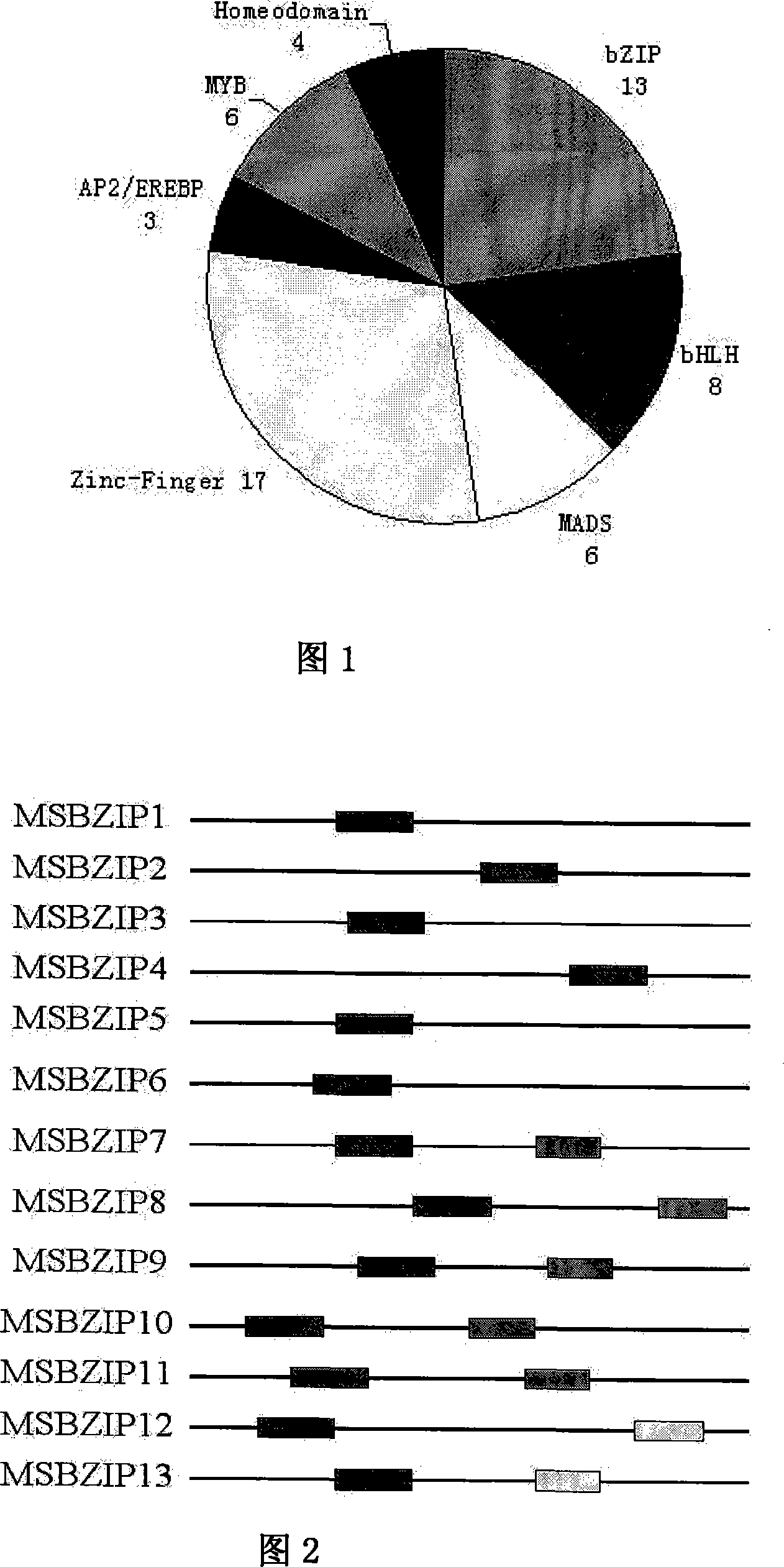
[0031] Use MEME analysis software to analyze the conserved segments of these 13 bZIP transcription factors, see Figure 2. They all contain a conservative alkaline leucine region, and MSBZIP7-12 except for the basic segment specific to bZIP protein , There is also a conservative segment with unknown function. According to the number and types of conserved segments, we divide these 13 transcription factors into 3 categories, among which MSBZIP1-6 belongs to the first category; MSBZIP7-11 belongs to the second category; and the third category includes MSBZIP12 and MSBZIP13.
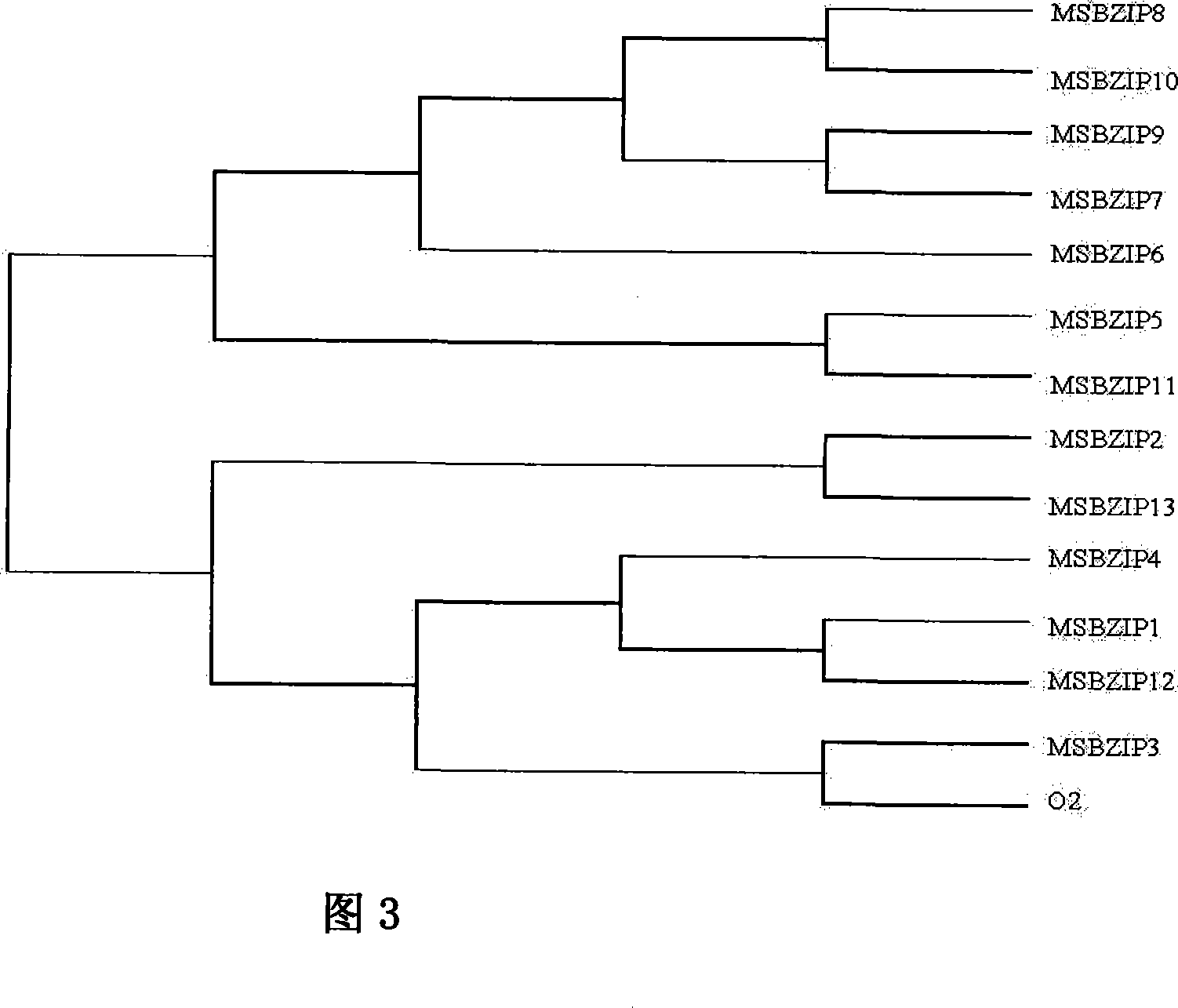
[0032] Use Megabase3.0 software to cluster these 13 bZIP transcription factors, see Figure 3. The results show that MSBZIP5-11 are relatively close in evolutionary distance, while MSBZIP1-4, MSBZIP12, MSBZIP13 and O2 are relatively close in distance. It may reveal two evolutionary patterns in these bZIP transcription factors.
Embodiment 3
[0033] Example 3: Expression profile analysis of bZIP transcription factor
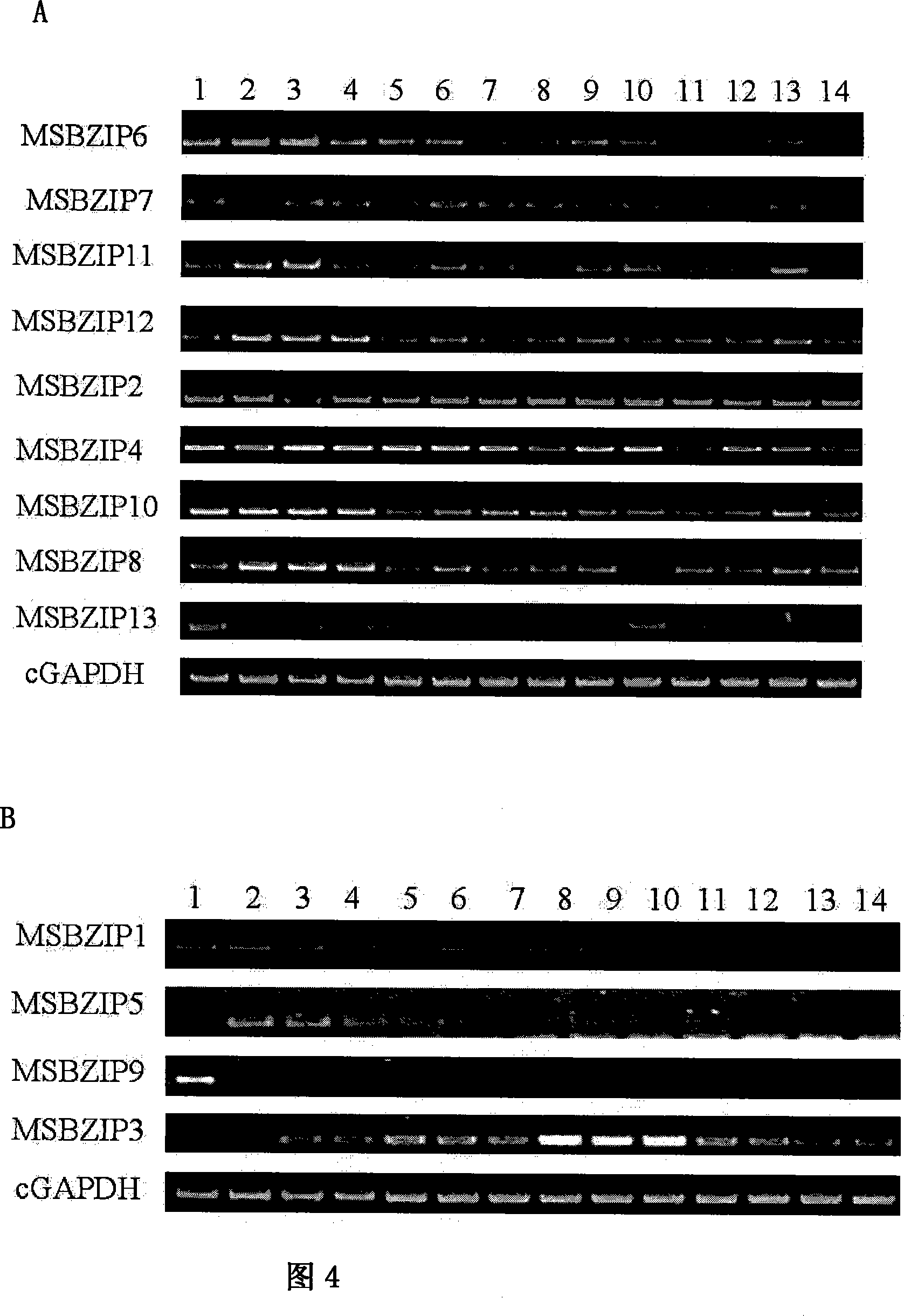
[0034] Using maize kernel RNA from 3-36 days after pollination as template, Oligo(dT) 16 Used as a primer for reverse transcription to obtain cDNA at various time points in the entire grain development stage. Primers were designed according to the sequences of these 13 transcription factors. See Table 1. Semi-quantitative PCR was performed using the cDNA at each time point as a template. The PCR products were analyzed by 1% agarose gel electrophoresis. See Figure 4, which proves that MSBZIP2, MSBZIP4, MSBZIP6, MSBZIP7, MSBZIP8, MSBZIP10, MSBZIP11, MSBZIP12, and MSBZIP13 are continuously expressed during grain development, see Figure 4-A; while MSBZIP1, MSBZIP3, MSBZIP5, MSBZIP9 are specifically expressed during development, see Figure 4 -B.
[0035] Table 1. Primer list of bZIP transcription factor
[0036] Gene name
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com