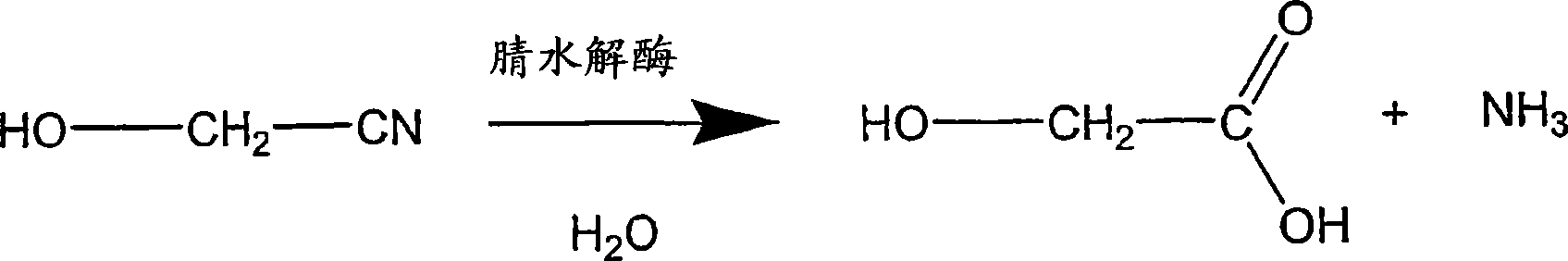
Enzymatic production of glycolic acid
A glycolic acid and glycolonitrile technology, applied in the field of microbiology and molecular biology, can solve the problem of no nitrilase activity and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0244] Construction of High Copy Nitrilase Expression Plasmid
[0245] Synthetic oligonucleotide primers
[0246] 165(5′-CGA CTGCAG TAAGGAGGAATAGGACATGGTTTCGTATAACAGCAAGTTC-3'; SEQ ID NO: 1) and
[0247] 166(5′-TGA TCTAGA GCTTGGAGAATAAAGGGGAAGACCAGAGATG-3′; SEQ ID NO: 2) (which incorporated PstI and XbaI restriction sites (underlined) respectively) was used for PCR amplification of genomic DNA from Acidovorax facilis 72W (ATCC 55746) (SEQ ID NO: 5) The nitrilase gene.
[0248] Typical PCR parameters are as follows:
[0249] Step 1: 5 minutes at 95°C
[0250] Step 2: 0.5 minutes at 95°C (denaturation)
[0251] Step 3: 0.5 minutes at 55°C (annealing)
[0252] Step 4: 1 minute at 74°C (extension)
[0253] Repeat steps 2-4 for 25 cycles.
[0254] PCR reagents were provided by Roche Diagnostics Corporation (Indianapolis, IN) and used as recommended.
[0255] The only change from the native A. facilis 72W sequence was a change in the first nucleotide from G to A to fac...
Embodiment 2
[0257] Expression of Active Nitrilase in E. coli
[0258] Plasmid pSW138 was used to transform E. coli MG1655 (ATCC 47076) and E. coli FM5 (ATCC 53911 ) to generate two strains, called (1) MG1655 / pSW138 and (2) FM5 / pSW138, respectively. Each strain was grown, induced, harvested and assayed for nitrilase activity (conversion of glycolonitrile to glycolic acid) as previously described. Each strain was repeated 6 times.
[0259] 1. Bacterial culture
[0260] The strain inoculum was grown in LB medium supplemented with ampicillin (50 mg / L) at 37°C with shaking (200 rpm) for 16-18 hours.
[0261] 2. Induction of nitrilase expression
[0262] Sufficient inoculum was added to fresh LB medium supplemented with ampicillin (50 mg / L) and IPTG (1 mM) to give a starting OD (600 nm) of approximately 0.1. The culture was incubated at 37°C with shaking (200 rpm) for about 6-8 hours.
[0263] 3. Bacteria harvest
[0264]Bacterial cells were harvested by centrifugation, removing a...
Embodiment 3
[0272] Construction of random mutagenesis library of nitrilase from Acidovorax facilis 72W by error-prone polymerase chain reaction
[0273] Genomic DNA was prepared from Acidovorax facilis 72W (ATCC 55746) using the Puregene(R) DNA Isolation Kit according to the manufacturer's instructions (Gentra Systems, Minneapolis, MN). According to the instructions provided by the GeneMorph(R) PCR Mutagenesis Kit (Stratagene, La Jolla, CA), the Acidovorax facilis 72W nitrilase gene (coding sequence; SEQ ID NO: 5) was subjected to error-prone PCR using the identifier SEQ ID Primers for NO: 3 (5'-GCGCATATGGTTTCGTATAACAGCAAGTTCC-3') and SEQ ID NO: 4 (5'-ATAGGATCCTTATGGCTACTTTGCTGGGACCG-3'). Use reaction conditions recommended to yield low mutation frequencies (0-3 mutations / kb) and intermediate mutation frequencies (3-7 mutations / kb). According to the instructions provided by the pTrcHis2 TOPO(R) TA Expression Kit (Invitrogen, Carlsbad, CA), 10% of the 1.1 kb PCR product was ligated into...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com