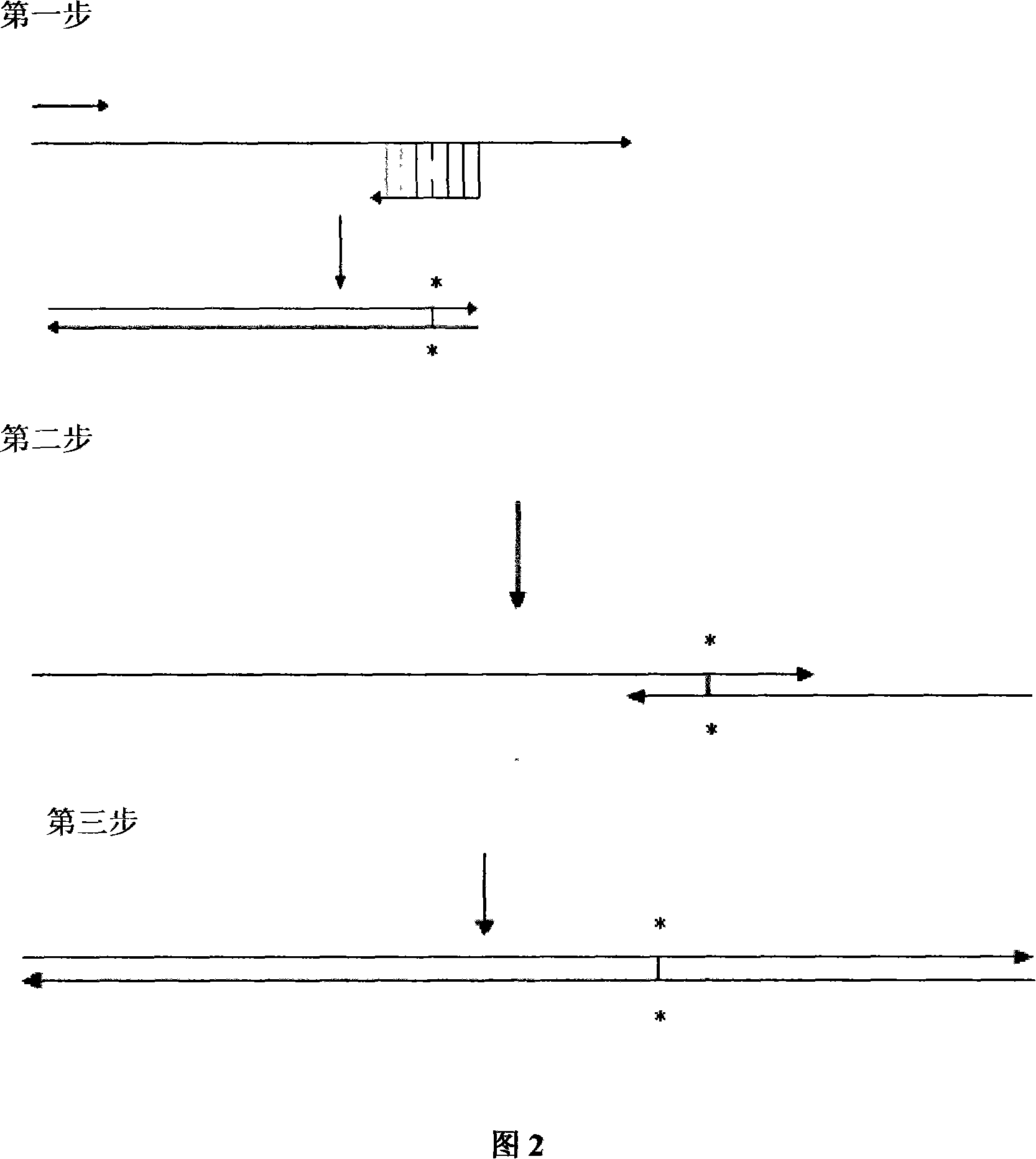Recombinant soluble human FGFR2 extracellular fragment and production method and application thereof
A technology of FGFR2 and production methods, applied in the direction of biochemical equipment and methods, applications, botany equipment and methods, etc., can solve problems such as carcinogenesis, cell transformation, and craniosynostosis, and achieve reduced levels, good activity, and production The effect of simple process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Construction of double mutants in the extracellular segment of FGFR2:
[0041] 1. Construction of the wild-type extracellular segment of FGFR2:
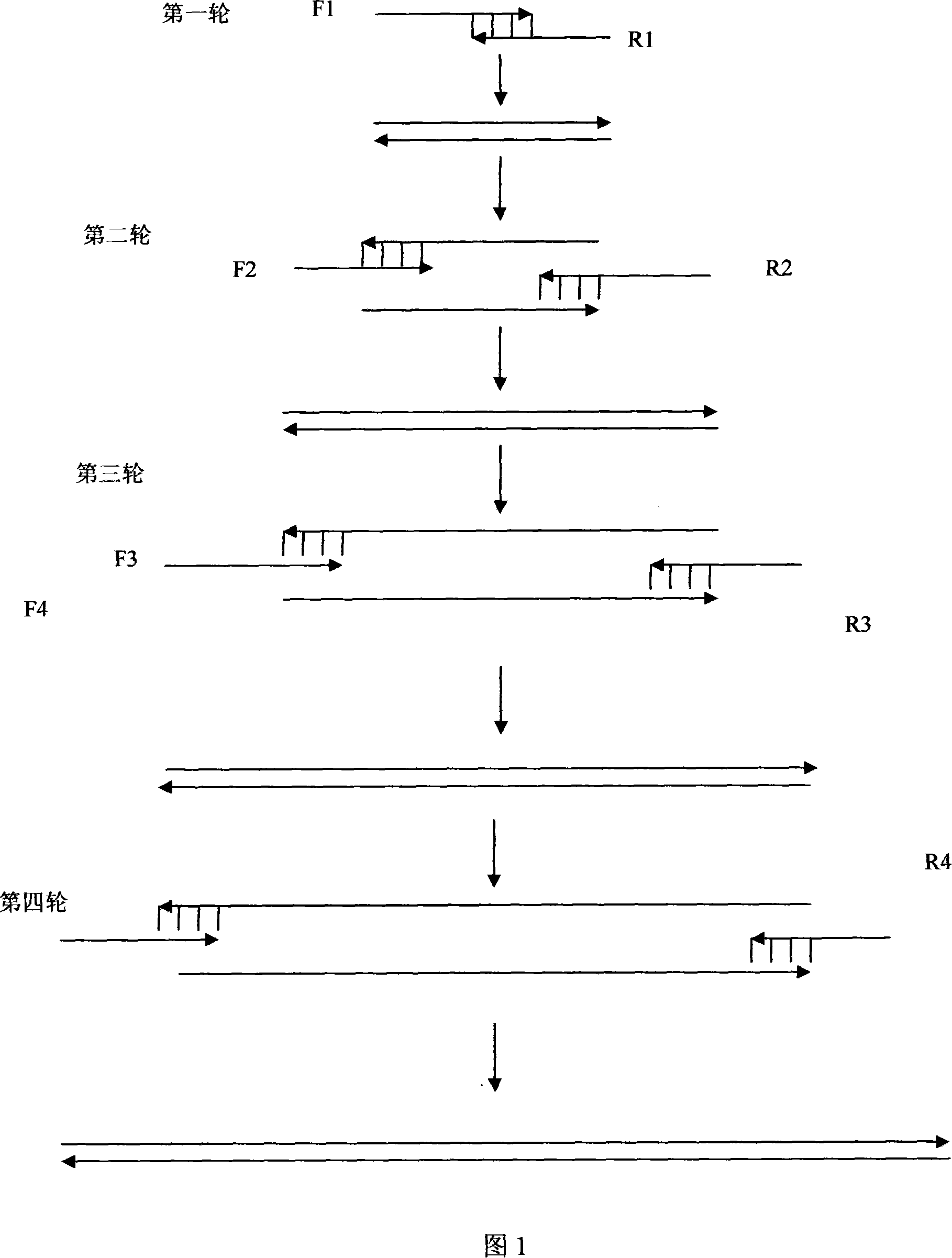
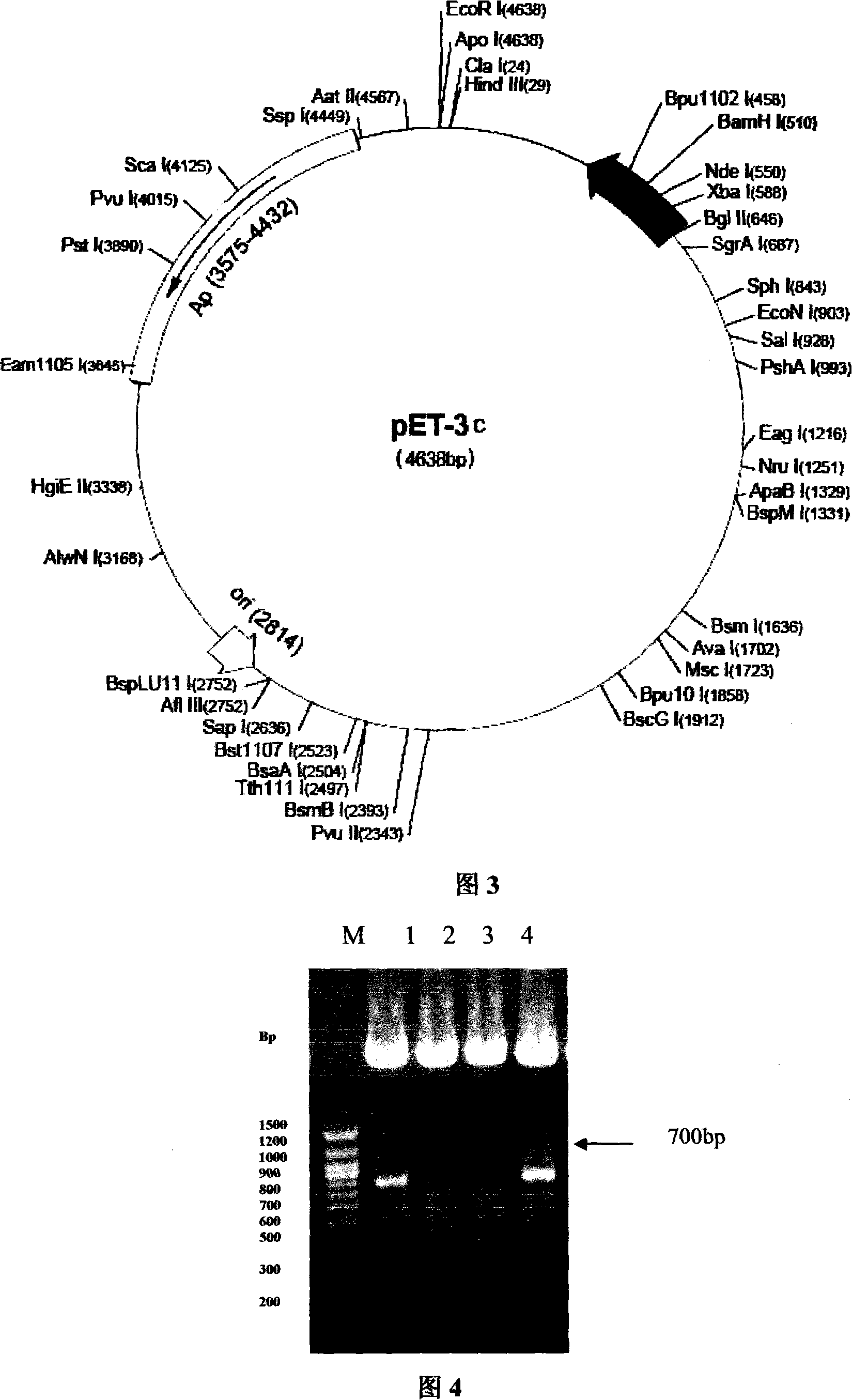
[0042]According to the amino acid sequence of the wild-type extracellular segment of the truncated FGF receptor, four pairs of primers were designed according to the codon preference principle of Escherichia coli, respectively marked as: upstream primers F1, F2, F3, F4; R4, as shown below, and Nde I and BamH I restriction endonuclease sites were introduced at the 5' ends of primers F4 and R4, respectively, to facilitate directional cloning of the gene. Each strip ranges from 105 to 106bp, and the full-length synthesis is 707bp. The reading frame contains a 684bp truncated version of the extracellular segment of FGFR2IIIC, starting from 150aa to 365aa, and adjacent to each strip. There is a 20bp overlap between the primers. There are four rounds of synthesis. In the first round, the innermost primers F1 and R1 are used for ov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com