Shikimic acid prepared bacterial strain and constructing method
A technology for producing strains and constructing methods, applied in the biological field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 , Construction of double knockout strain W3110 (ΔaroLΔaroK)
[0028] 1.1 Knockout of aroL and aroK genes in BW25113 (E.coli Genetic Stock Center)
[0029] With reference to the literature of Gust.B. et al. (PCR-targeting system in Streptomyces coelicolor, Gust.B., KieserT.et al, 2002), the PCR-Targeting method was used to knock out the aroL and aroK genes in the Escherichia coli genome, as follows :
[0030] 1.1.1. Obtainment of single knockout strain BW25113 (ΔaroL)
[0031] Using the plasmid pIJ773 (E.coli Genetic Stock Center) as a template, the aac (Apra) gene in the plasmid pIJ773 was amplified by PCR using primers aroK-up and aroK-down. The primer sequences are as follows:
[0032] aroK-up: 5'-tacgctaatcttacccggtgattatcgccagagcggtgattccggggatccgtcgacc-3';
[0033] aroK-down: 5'-tactgtacccgcagacgagtgtatataaagccagaattatgtaggctggagctgcttc-3';
[0034] The amplification conditions are: 94°C, 2min, 94°C, 45sec, 55°C, 45sec, 72°C, 90sec, 30 cycles.
[00...
Embodiment 2
[0053] Example 2 1. Construction of recombinant plasmid pSUFEBPT
[0054] 2.1. Cloning of recombinant plasmid pSU-F
[0055] Using the genome of E.coli DH5α (Amersham Company) as a template, use primers P148L(+), P148L(-), aroFSac I(+) and aroFSac I(-) to perform overlapping PCR to amplify the aroF gene of E.coli DH5α , the primer sequences are as follows:
[0056] P148L(+): 5'-ttagatctgaatagcccgcaatacctgggc-3';
[0057] P148L(-): 5'-gctattcagatctaacgcttccgtcgccagtgg-3';
[0058] aroFSac I(+): 5'-aacgagctcaccggaaagtcctcgggcataag-3';
[0059] aroFSac I(-): 5'-aacgagctccgacttcatcaatttgatcgcgtaa-3';
[0060] First, a fragment was amplified with the primer pair aroFSac I(+) and P148L(+), the amplification conditions were: 94°C, 2min, 94°C, 45sec, 56°C, 45sec, 72°C, 1min, 30 cycles; The primer pair aroFSac I(-) and P148L(-) amplified another fragment. The amplification conditions were: 94°C, 2min, 94°C, 45sec, 56°C, 1min, 72°C, 1min, 30 cycles; gel recovery as above Two PCR...
Embodiment 3
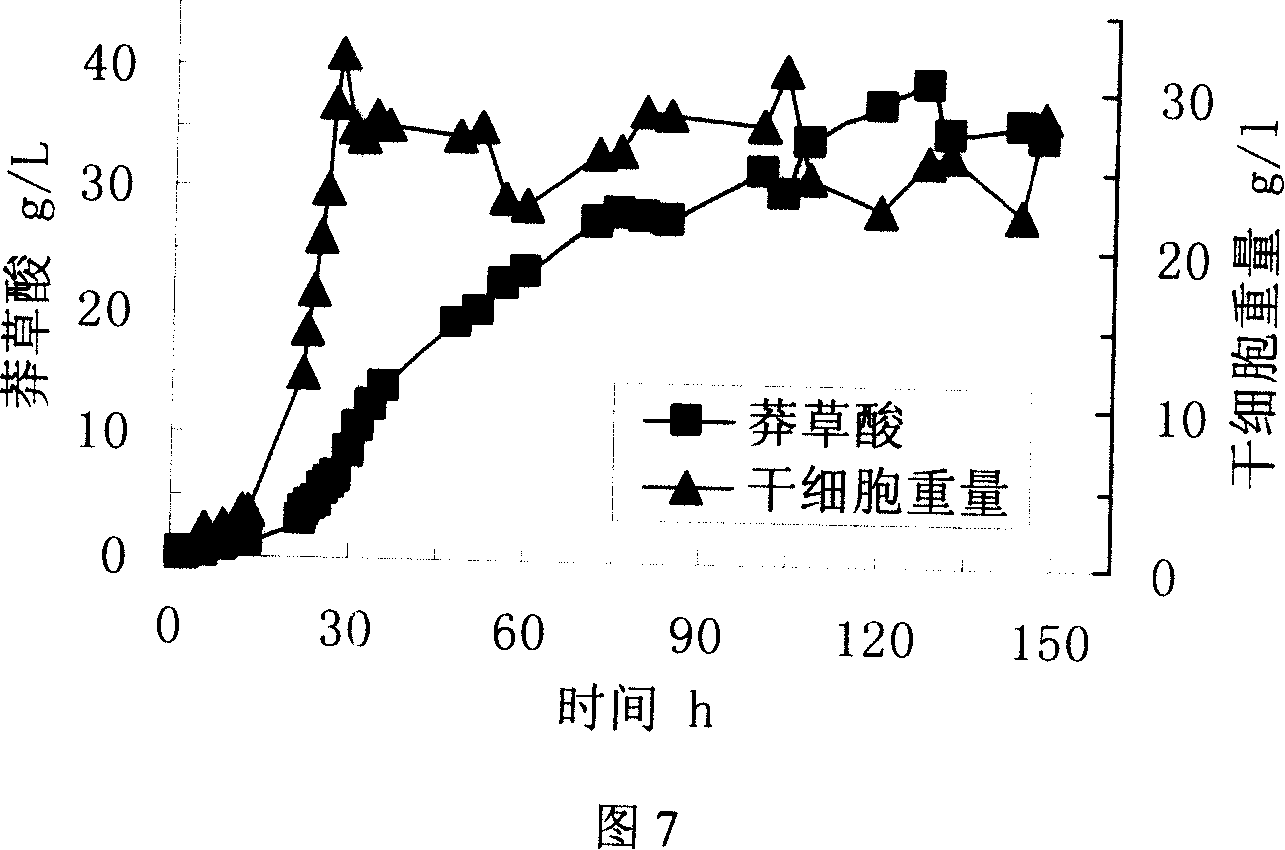
[0096] Example 3 , Obtaining of shikimic acid expression strain
[0097] The Escherichia coli host bacterium W3110 (ΔaroLΔaroK) obtained in Example 1 that knocked out the aroL and aroK genes was made into competent cells, and then the recombinant expression plasmid pSUFEBPT obtained in Example 2 was transferred into competent cells by chemical transformation (specific method Referring to molecular cloning III), Escherichia coli host strain W3110 (ΔaroLΔaroK) expressing shikimic acid was obtained.
[0098] The shikimic acid expression strain W3110 (ΔaroLΔaroK) obtained above was submitted to the China Center for Type Culture Collection (CCTCC) located in Wuhan on September 4, 2006 for deposit, and the deposit number is CCTCC M206083.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com